Figures & data
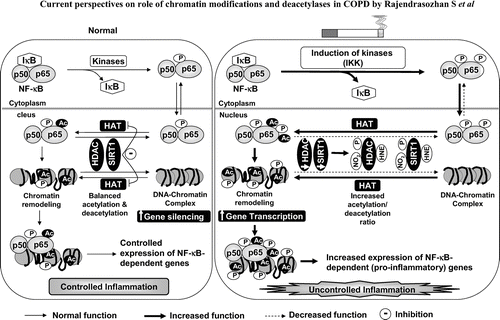
Figure 1 Regulation of NF-κ B-dependent gene expression by histone acetyltransferases (HATs) and histone deacetylases (HDACs). DNA is coiled around the histone proteins and form chromatin structure. Acetylation of histones leads to opening of the chromatin and increasing the accessibility of transcription factors, such as NF-κ B. HATs acetylate histones and RelA/p65 (subunit of NF-κ B) which facilitate the binding RelA/p65-p50 heterodimer onto the pro-inflammatory gene promoters and thereby increasing gene transcription. Deacetylation results in DNA rewinding around histone proteins, and decreasing gene transcription. Thus, HDACs are involved in the maintenance of histone acetylation and deacetylation balance thereby control the transcription of gene. In response to cigarette smoke exposure, deacetylases (especially HDAC2 and SIRT1) are post-translationally modified by phosphorylation, carbonylation, aldehyde adducts formation and/or nitration/nitrosylation leading to increased ubiquitination and subsequent degradation of deacetylases. Decrease in deacetylases results in increased histone acetylation/deacetylation ratio resulting in increased acetylation of histones (chromatin remodeling) and NF-κ B subunits leading to increased transcription of pro-inflammatory mediators. Dotted lines indicate less function (less phosphorylation and acetylation) whereas solid lines indicate increased function (more phosphorylation and acetylation). HAT: Histone acetyltransferase; HDAC: Histone deacetylase; P: Phosphorylation; Ac: Acetylation; 4-HNE: 4-hydroxy-2-nonenal; NO2: Nitrogen dioxide.