Figures & data
Figure 1. Summary of the chimerism results before and after three DLIs into two stable mixed chimeric recipients. The y-axis shows the percentage of donor chimerism in the granulocyte fraction for H550 (A) and H552 (B), and PBMC fraction for H550 (C) and H552 (D). The x-axis shows times in months. Baseline reported chimerism values for each recipient for the 8 months preceding the first DLI with the last time point taken two weeks prior to the first DLI. The rest of the chimerism values are reported following each DLI with zero representing the time of the DLI.
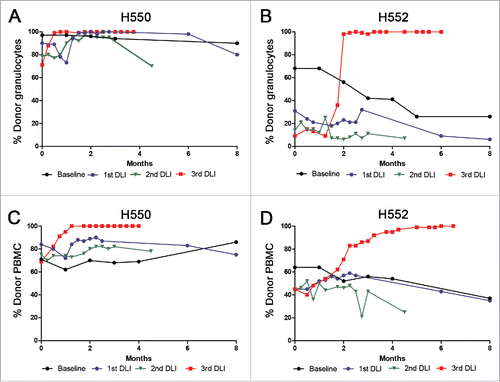
Table 1. Summary of the donor sensitization attempts followed by DLI into the mixed chimeric recipient.
Figure 2. Variable number tandem repeat (VNTR) assay. Genomic DNA of H555 and H552 (A) H554 and H550 (C) were isolated and amplified by PCR for VNTR. The data are represented by peaks with the shaded areas representing area under the curve (AUC). The top panel shows the marrow donor VNTR prior to any cell injections from the mixed chimera. The middle panel shows marrow donor VNTR one month after the last set of cell injections from the mixed chimera. The bottom panel shows the VNTR of the transplanted recipient prior to haematopoietic cell transplant as a comparison. Titrated mixtures of pre-transplant donor and recipient DNA shown for H555 and H552 (B) and H554 and H550 (D) were shown in a logarithmic scale to determine the limits of detection.
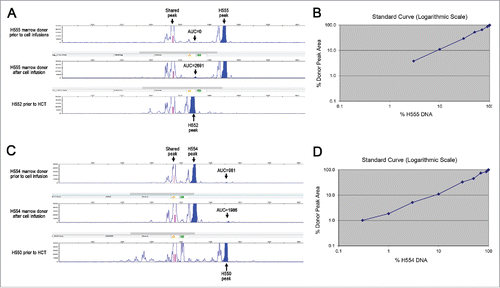
Figure 3. Fluorescence-activated cell sorting of peripheral blood dendritic cells. The first gate utilized forward scatter and side scatter properties to eliminate cell debris (left plot). The second gate was then used to exclude CD14+, DM5- monocytes and the DM5+, CD14- granulocytes from the sorted population (middle plot). MHC class II+ cells determined using an antibody against DLA-DR, and canine CD11c+ cells were then sorted for use in chimerism analysis or in MLRs (right plot).
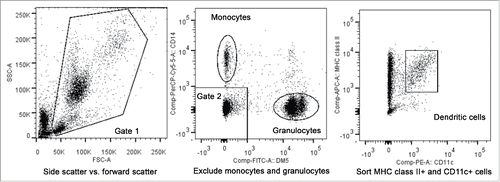
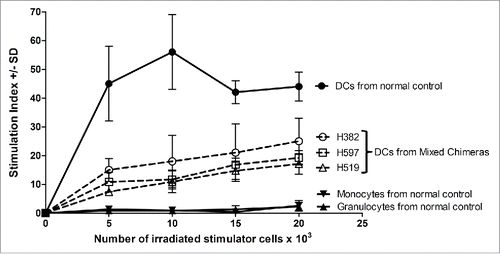
Figure 4. Functional characterization of sorted cell populations assayed in MLR. The monocyte, granulocyte, and peripheral blood DCs populations were sorted as shown for and used as stimulator cells. Constant numbers of third party responding PBMC (1×105 cells per well) were mixed with varying numbers of irradiated stimulator cells (5–20×103). The responders and stimulators were MHC-mismatched. The stimulation index was calculated as the average counts per minute of the responder cells with stimulator cells added divided by the average counts per minute of responder cells only plus or minus the standard deviation (SD). The solid line and filled in data points were for the normal control, while the three samples from mixed chimeric recipients H382, H597, and H519 were shown with dotted lines and open data points.