Figures & data
Figure 1. Presentation of the EV-TRACK study summary add-on.
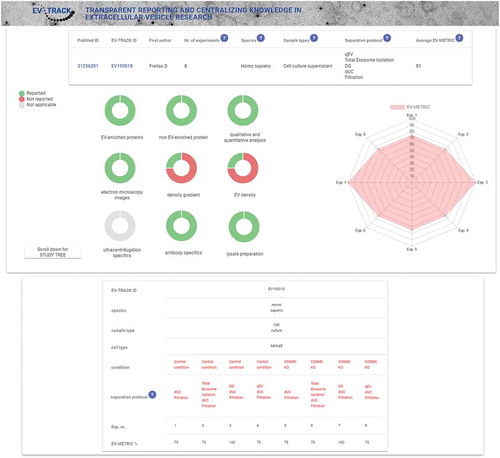
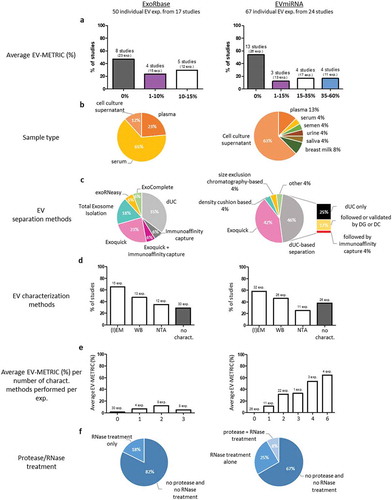
Figure 2. EV-TRACK analysis of ExoRbase and EVmiRNA publications.
Data availability statement
All data that were collected during the course of this study, and that support its findings are available online http://evtrack.org.
