Figures & data
Figure 1. Computational disorder analysis of curli related proteins. Uversky plots of mean net charge against mean hydrophobicity for proteins involved in curly assembly. (A) CsgA (B) CsgB, (C) CsgC, (D) CsgE, (E) CsgF, and (F) CsgG. Except for CsgC and CsgG, both of which fall within the ordered protein region (black diamond in panels C and F), the values for CsgA, CsgB, CsgE, and CsgF (black diamond in panels A, B, D, and E) fall near the order-disorder boundary (represented by the solid line) suggesting that these could be disordered proteins. The location of a set of ordered (blue), and disordered (red) proteins are shown for comparison (from Ponder.com).
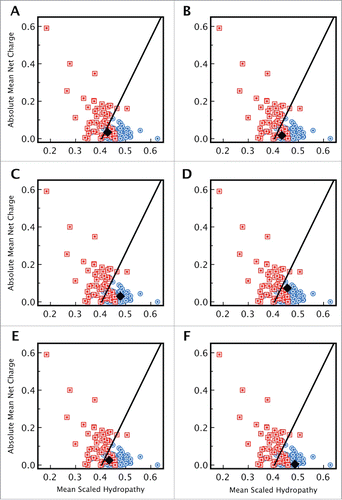
Figure 2. Distribution of disorder and secondary structure within CsgE and CsgF. Sequences of (A) CsgE and (B) CsgF indicating the regions of disorder predicted by PONDR (regions highlighted in red). PSIPred predicted regions of secondary structure are represented using purple rectangles (α-helix) and yellow arrows (β-sheet). Sequence analysis indicates that for CsgE approximately 34% of the residues are in an α-helix, 22% in a β-sheet, and 43% in a random coil, while for CsgF approximately 28 % of the residues are in an α-helix, 27 % are in a β-sheet, with the remainder (45%) being unstructured. The 19 residue signal sequence for each protein is shown in light gray.
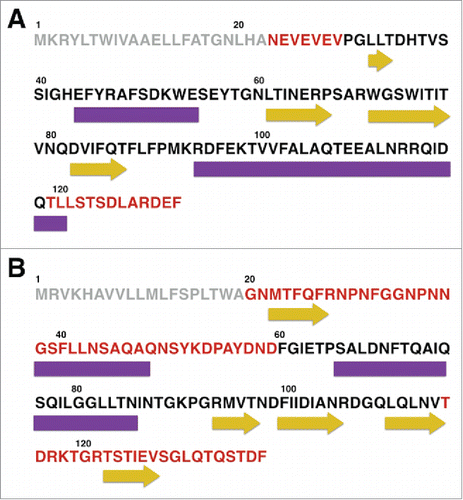
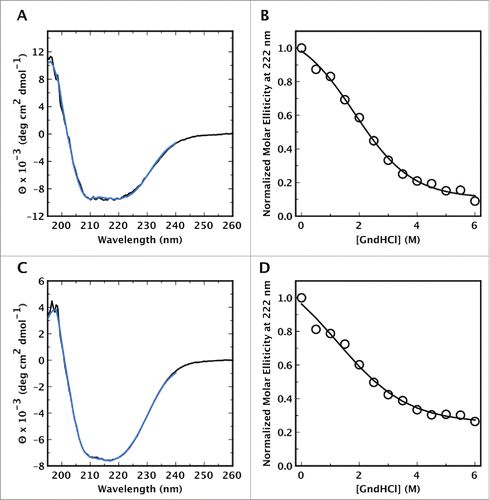
Figure 3. Circular dichroism spectra of CsgE, and CsgF, and their unfolding with GndHCl. The CD spectra of (A) CsgE and (C) CsgF in solution (black line) are indicative of a mixed distribution of secondary structures, with no distinct characteristic expected for a predominantly α-helical or predominantly β-sheet structure. Spectra were deconvoluted using DICHROWEB and CsgE was estimated to contain a distribution of 28% α-helix, 23% β-sheet and 49% random coil structure. In the case of CsgF the content of α-helix, β-sheet, and coil structure was found to be 28 %, 15 %, and 57 % respectively. The fitted spectra obtained via DICHROWEB are shown in blue. Both (B) CsgE and (D) CsgF exhibited weakly cooperative, or noncooperative unfolding curves with a midpoint denaturation of approximately 2 M GndHCl.