Figures & data
Figure 2. The prokaryotic community composition of Sargassum waste at phylum level. LB, Long Beach; HS, Harrismith beach. The DNA extracted from quadruplicate Sargassum samples from each beach was pooled into one representative sample, and then used to prepare the Illumina sequencing libraries targeting the V3-V4 region of the 16S rRNA gene.
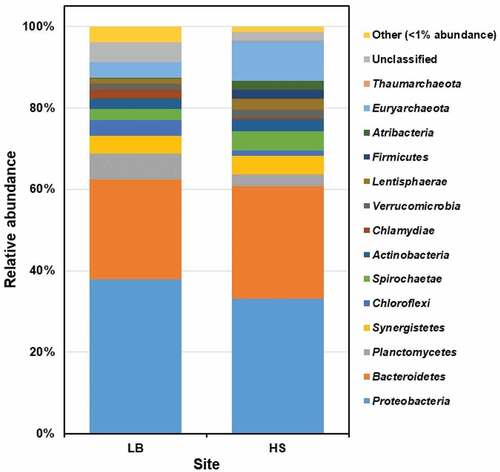
Figure 3. The prokaryotic community composition of Sargassum waste at family level. LB, Long Beach; HS, Harrismith beach. The DNA extracted from quadruplicate Sargassum samples from each beach was pooled into one representative sample, and then used to prepare the Illumina sequencing libraries targeting the V3-V4 region of the 16S rRNA gene.
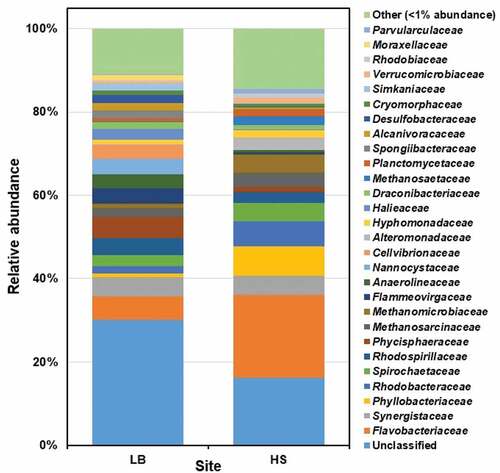
Figure 4. The prokaryotic community composition of Sargassum waste at genus level. LB, Long Beach; HS, Harrismith beach. The DNA extracted from quadruplicate Sargassum samples from each beach was pooled into one representative sample, and then used to prepare the Illumina sequencing libraries targeting the V3-V4 region of the 16S rRNA gene.
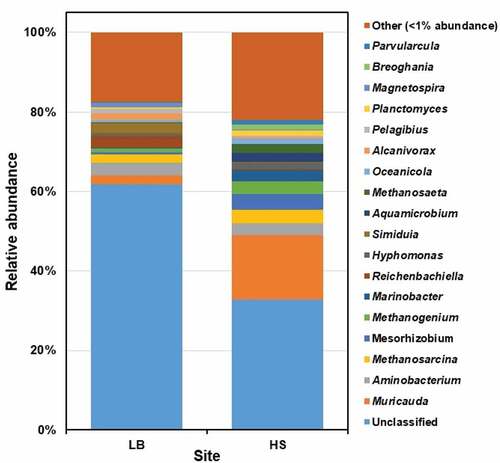
Figure 5. The fungal community composition of Sargassum waste at phylum level. LB, Long Beach; HS, Harrismith beach. The DNA extracted from quadruplicate Sargassum samples from each beach was pooled into one representative sample and then used to prepare the Illumina sequencing libraries targeting the ITS2 region of the rRNA.
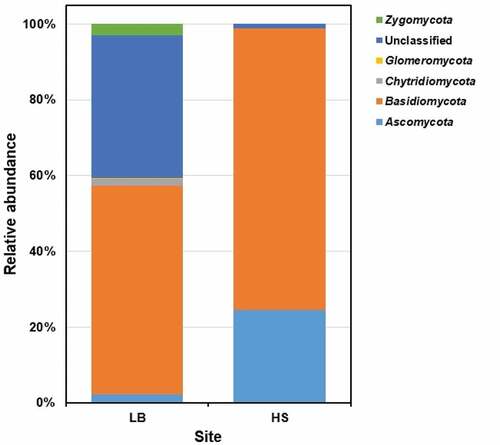
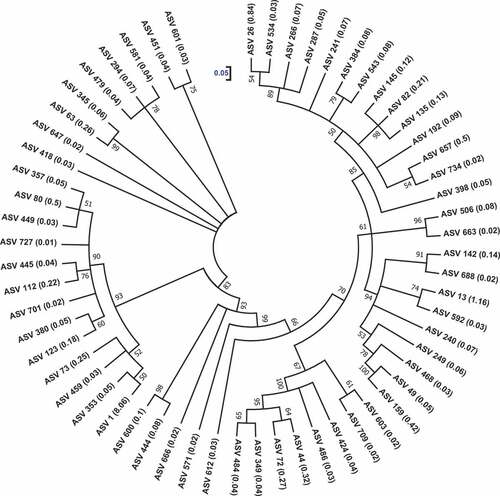
Figure 6. The phylogenetic relatedness of alginate lyases-producing prokaryotic communities associated with Sargassum waste. ASV, amplicon sequence variant. The cladogram was drawn using maximum-likelihood algorithm. The bootstrap percentage values of 1000 replications are specified at the nodes. The scale denotes 0.05 substitutions per nucleotide position. Numbers inside the parentheses denote the relative abundance (%) of the ASVs.