Figures & data
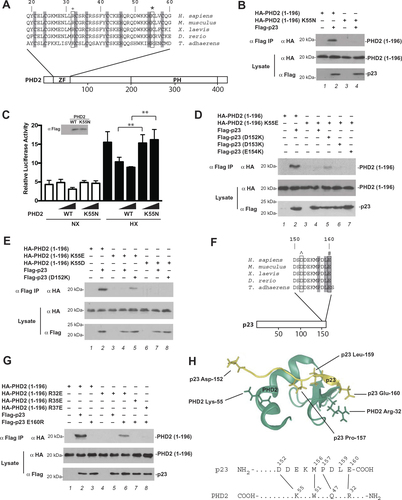
Figure 1 Functional characterization of K55N PHD2. (A) Diagram of PHD2, showing the location of zinc finger (ZF) and prolyl hydroxylase (PH) domains. Sequence of zinc finger across various metazoan species is shown at top. *= Lys-55 and += Arg-32 (human nomenclature). Shading = zinc chelating residues. (B, D, E, and G) HEK293FT cells were transfected with constructs for the indicated proteins. Cells were lysed, the Flag-tagged proteins were immunoprecipitated, and the immunoprecipitates examined for the absence or presence of HA-tagged PHD2 (1–196) by anti-HA Western blotting. Anti-HA and anti-Flag Western blots of lysates are also shown. Positions of molecular weight markers were as indicated. (C) HEK293FT cells in 96-well plates were transfected using Lipofectamine 2000 with 8 ng of (eHRE)3-Luc, 8 ng of RL-TK (which expresses Renilla luciferase under the control of the HSV thymidine kinase promoter), and either 0.8 or 2.5 ng of either pcDNA3-Flag-PHD2 or pcDNA3-Flag-PHD2 K55N. DNA doses were held constant by the addition of pcDNA3. Eight hr after transfection, cells were exposed to 1% O2 (HX) or maintained under normoxia (NX) for an additional 16 hr. All cells were lysed, and luciferase activities were measured and normalized to that of the Renilla luciferase internal transfection control. Shown are means ± SD, n = 3. **, p < 0.01 by student’s t-test. Anti-Flag Western blot of lysates of HEK293FT cells transfected with expression constructs for WT or K55N PHD2 is also shown. (F) Diagram of p23. ^= Asp-152 and # = Glu-160 (human nomenclature). Shading = P, L, and E of PXLE motif. (H) Top: model of p23 (152–160), which contains a PXLE motif, bound to the zinc finger of PHD2 (residues 20–59) generated as previously describedCitation12 and visualized using Protean 3 (DNASTAR). PHD2 is shown in green, p23 in yellow. Side chains of PHD2 Lys-55 and Arg-32, and p23 Asp-152, Pro-157, Leu-159, and Glu-160 are shown. Bottom: proposed contacts between residues in the C-terminal tail of p23 and residues in zinc finger of PHD2. N- and C-termini of the proteins are denoted by NH2 and COOH, respectively.