Figures & data
Figure 1 Structures of enveloped, DNA oncolytic viruses in complex with their cellular receptors. (A) Schematic diagram of herpes simplex virus-1 (HSV-1). (B) HSV-1 utilizes its surface exposed glycoprotein D ectodomain to bind host cellular receptor herpes virus entry mediator A (HveA) ectodomain (PDB: 1JMA). (C) Glycoprotein D of HSV also interacts with the first Ig domain of nectin 1 at 1:1 stoichiometry. Nectin-1 binding site on gD differs from HveA binding site, as evident from the crystal structures arranged in the same orientations (PDB: 3SKU).
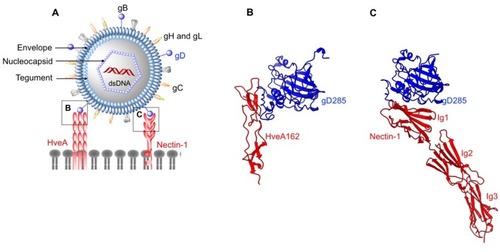
Figure 2 Structures of enveloped, RNA oncolytic viruses in complex with their cellular receptors. (A) Schematic diagram of vesicular stomatitis virus. (B and C) Vesicular stomatitis virus (VSV) surface glycoproteins (VSV-G) identify and interact with cysteine-rich domains (CRD) on low-density lipoprotein receptors (LDLR) expressed in cancer cells. Different CRDs interact with VSV-G at identical locations as evident from crystal structures arranged in the same orientation (PDB: 5OLY and 5OY9). (D) Schematic diagram of Newcastle disease virus (NDV). (E) Newcastle disease virus (NDV) surface protein hemagglutinin-neuraminidase (HN) exploits cell surface sialic acid (SA) as the cellular receptors. Two SA binding sites exist on HN dimers, SA1, and SA2 (PDB: 1USR). (F) Schematic diagram of Measles virus (MV). (G and H) measles virus (MV) H binds CD46 short consensus repeats (SCR) 1, SCR2, SCR1-2 interface (PDB: 3INB) and domain 1 of nectin-4 (PDB: 4GJT).
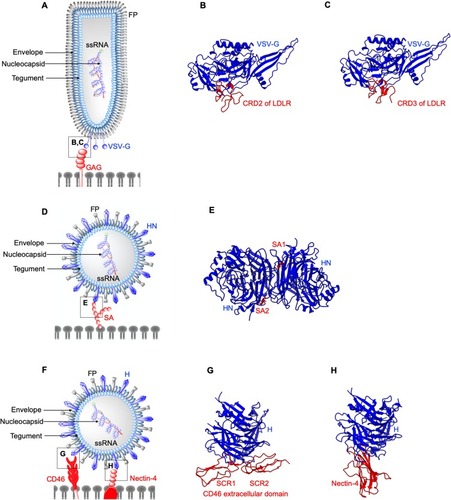
Figure 3 Structures of non-enveloped, DNA oncolytic viruses in complex with their cellular receptors. (A) Schematic diagram of human adenovirus (AdV). (B) Coxsackievirus-adenovirus receptor (CAR) extracellular D1 domain interacts with a monomer of AdV12 knob (PDB: 1P69). (C) AdV 11 exploits CD46 as the primary receptor. AdV knob monomer interacts with short consensus repeat (SCR) 1 and SCR1-2 interface. Another knob monomer binds the base of SCR2 (PDB: 3O8E). (D) AdV52 utilizes its short fibers to bind polysialic acid (polySA) and monomer 1 and 3 of the knob trimers interact with two polySA (PDB: 6G47). (E) Desmoglein 2 (DSG2) acts as the receptor for AdV3 with two distinct receptor:knob ratios of 1:1 and 1:2 observed. DSG2 EC2 and EC3 interact with monomer 1 and 2 of AdV3 knob, respectively (PDB: 6QNT). (F) Schematic diagram of human Reovirus (RV). (G) RV exploits cell surface sialic (SA) acid as its attachment receptor to tether the virus in close proximity to host membrane to interact with an entry receptor, junction adhesion molecule-A (JAM-A). SA binds to the stalk of the trimeric sigma protein (PDB: 3S6X), whereas (H) JAM-A D1 domain interacts with the head of the trimeric sigma protein (PDB: 3EOY).
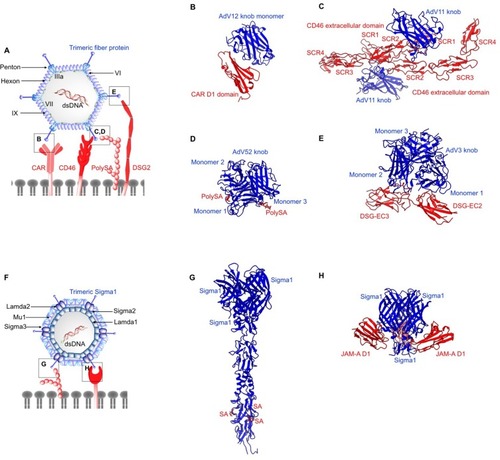
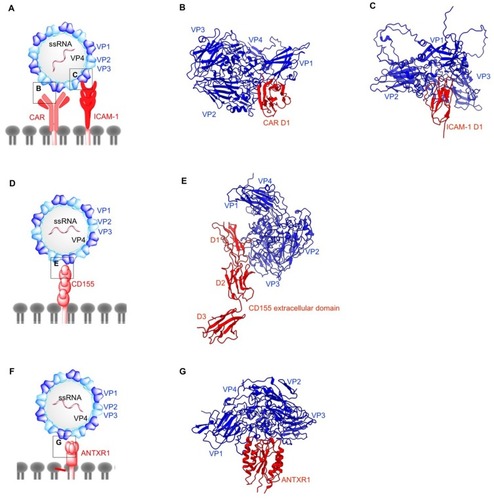
Figure 4 Structures of non-enveloped, RNA oncolytic viruses in complex with their cellular receptors. (A) Schematic diagram of coxsackievirus (CV). (B) D1 domain of coxsackievirus-adenovirus receptor (CAR) acts as the binding site for coxsackievirus B (CVB) capsid proteins VP1-VP3 (PDB: 1JEW). (C) Coxsackievirus A variant 24 (CVA24v) capsid proteins VP1 and VP2 interact with the D1 domain of intracellular adhesion molecule-1 (ICAM-1) (PDB: 6EIT). (D) Schematic diagram of poliovirus (PV). (E) Poliovirus utilizes CD155 on the cell surface as its cellular receptor. Similar to ICAM1 and CAR, CD155 D1 domain binds PV capsid proteins VP1 and VP2 from one protomer and VP3 from the adjacent protomer (PDB: 3J8F). (F) Schematic diagram of Seneca Valley Virus (SVV). (G) Anthrax toxin receptor 1 binds to surface-exposed loops of VP1–VP3 on SVV capsid (PDB: 6CX1).