Figures & data
Table I. Primer sequences used for PCR amplification of individual exons of PHF6.
Figure 1. Representative SSCP and DNA sequencing of PHF6 gene in the cancers. SSCP (A–C) and DNA sequencing analysis (D–F) of PHF6 gene from a T-ALL, an AML and a HCC in cancer (Lane T) and normal tissues (Lane N). A–C. The arrows in lane T indicate aberrant bands as compared with SSCP of normal tissues (lane N). D–F. DNA sequencing of the aberrant SSCP bands in A, B and C show four base insertions in D (c.90_91insCCCG), one base duplication in E (c.27dupA) and a substitution of a base in F (c.673C > T), respectively.
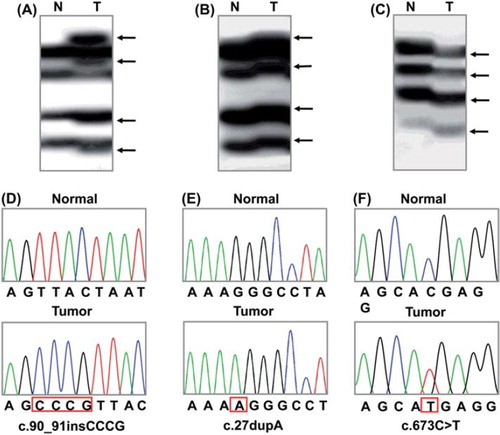
Table II. Summary of PHF6 mutations.