ABSTRACT
Trophic interactions between threatened species complicate management. Similarly, interactions between threatened species and pest species present management challenges, given that pest control can lead to non-target impacts (e.g. trophic cascades or secondary poisoning). There are records of the critically endangered Norfolk Island Morepork Ninox novaeseelandiae undulata consuming both threatened songbirds and invasive rodents that are subject to management interventions. Nevertheless, the diet of the morepork remains largely unknown. We visually screened regurgitated pellets using a microscope, alongside environmental DNA (eDNA) screening of pellets and scats, to investigate the diet of the Norfolk Island Morepork. A total of 113 pellets and 19 scats were collected between October 2020 and June 2021. All moreporks screened with eDNA metabarcoding had consumed invasive rodents and at least one-third of samples contained rodents. The owls were also found to have consumed four of five endemic songbirds and possibly an endemic parrot, most of which are threatened. Environmental DNA metabarcoding detected more taxa overall, but visual screening identified a greater richness of Orthoptera and Coleoptera in the diet. The frequency with which the Norfolk Island Morepork consumed rodents presents a conundrum for conservation managers. Control of invasive rodents is considered essential to support threatened songbirds, yet this same action places the species at risk of secondary poisoning. Urgent investigations are needed to identify effective control methods for invasive rodents that are safe for non-target species.
Introduction
Understanding trophic interactions can be fundamental to species conservation and ecosystem management (Soulé et al. Citation2003; Sousa et al. Citation2019). At a time of biodiversity crisis, as the number of threatened species increases, trophic interactions that include species of conservation concern present increasing challenges for managers (Roemer and Wayne Citation2003; Canale and Bernardo Citation2016). Island systems with a globally disproportionate number of threatened species, alongside simplified food webs and limited dietary redundancy, exemplify these challenges (Whittaker and Fernández-Palacios Citation2007).
The presence of invasive rodents is a leading cause of biodiversity loss, and adds further to the complexity of trophic interactions (Howald et al. Citation2007). Invasive rodents are typically widespread dietary generalists and have been implicated in the decline and extinction of many plant and animal species (St Clair Citation2011; Doherty et al. Citation2016; Russell et al. Citation2017). Programmes to control or eradicate rodents are therefore common practice (Keitt et al. Citation2011). When successful, these rodent management programmes can have substantial benefits to species and ecosystem recovery (Croll et al. Citation2016; Jones et al. Citation2016).
The management of invasive rodents routinely involves the use of anticoagulant rodenticides (Fisher et al. Citation2019) and this approach has often been very successful (Howald et al. Citation2007; Wheeler et al. Citation2019). However, because of evolved resistance in rodents, the rodenticides have by necessity become increasingly toxic with longer latency periods (Hadler and Buckle Citation1992; Marquez et al. Citation2019). Anticoagulant rodenticides are now characterised as either first- or second-generation toxins. Both forms prevent blood clotting and cause vertebrate death through haemorrhaging (Park et al. Citation1984). Second-generation baits are now 100–1000 times more toxic and have longer biological half-lives than first-generation baits (Huckle et al. Citation1988). Second-generation baits are intended to kill a rodent with a single dose, as opposed to first-generation baits which typically require multiple feeds. Consequently, the toxins from second-generation baits remain in animal tissue for longer and are less likely to be entirely metabolised by rodents before death (Huckle et al. Citation1988; Erickson and Urban Citation2004). When a rodent that has ingested anticoagulant bait is consumed by a predator or scavenger, any secondary poisoning that occurs invariably has physiological consequences and is often fatal (Lohr and Davis Citation2018). Because of the markedly higher toxicity of second-generation baits and their extended environmental latency, the use of these baits increases both the prevalence of secondary poisoning and the mortality rate in non-target wildlife, compared to the effects of first-generation baits (Erickson and Urban Citation2004; Van den Brink et al. Citation2018).
Birds of prey are particularly susceptible to secondary poisoning since rodents and other small mammals often comprise a substantial proportion of their diet. This is especially true when second-generation baits are involved. In settings where secondary poisoning is known or suspected to be occurring, a comprehensive assessment of diet can be informative for conservation managers. The traditional method to quantify the diet of birds of prey is to undertake a visual analysis of regurgitated prey remains (pellets) (Maser and Brodie Citation1966; Cooke et al. Citation2006). With the advent of environmental DNA (eDNA) techniques, traces of DNA sourced from the environment rather than directly from focal species can now also be used to detect the presence of organisms under a range of settings, including dietary screening (Cavallo et al. Citation2018; Quasim et al. Citation2018; Menning et al. Citation2023). eDNA techniques are especially well suited to establishing the dietary composition of birds of prey, given that they pass both pellets and faeces, and frequently do so at established perching or roosting sites (Driver Citation1949).
The Norfolk Island Morepork Ninox novaeseelandiae undulata is a critically endangered owl restricted to Norfolk Island, with a population estimated at 25–35 individuals (Threatened Species Scientific Committee Citation2016; F Sperring, unpubl. data). After a genetic rescue in 1987, where the last remaining female N. n. undulata paired and successfully bred with a male N. n. novaeseelandiae (New Zealand Morepork), this is now recognised as a hybrid population. The known diet of the Norfolk Island Morepork is based on a single pellet, prey remains from one nest-box, and incidental observations (Olsen Citation1997). These data suggest that invertebrates are a prominent prey type, though vertebrates probably predominate in terms of prey biomass (Olsen Citation1996). The White Tern Gygis alba, at least two threatened songbirds (Norfolk Island Robin Petroica multicolor and Slender-billed White-eye Zosterops tenuirostris), and the introduced Polynesian Rat Rattus exulans have been documented as vertebrate prey (Olsen Citation1996).
On Norfolk Island, the invasive Black Rat Rattus rattus and Polynesian Rat are implicated in the decline and extinction of many threatened flora and fauna (Nance et al. Citation2021, Citation2023). Invasive rodents are currently controlled intensively within protected areas to suppress their numbers. Rodent control programmes using toxic baits have been maintained since the 1990s, commencing with the use of first-generation baits, transitioning to second-generation baits in 2011. The control of rodents with toxic baits may pose a direct threat to the Norfolk Island Morepork population through secondary poisoning. In recognition of this as yet unquantified threat, bait deployment during the morepork breeding season has been modified to use first-generation and non-anticoagulant baits since 2015 (Nance et al. Citation2023). While second-generation baits remain in use throughout the remainder of the year to protect threatened songbirds, the quantity and toxicity have been reduced.
Our aim in this study was to quantify the diet of the Norfolk Island Morepork to inform key conservation management actions. We utilised visual analysis of pellets, and eDNA metabarcoding of pellets and scats, to identify prey items and quantify the dietary breadth of the Norfolk Island Morepork.
Methods
Study site
Norfolk Island is a small oceanic island (3460 ha) in the SW Pacific. The island has largely been cleared of native vegetation, with the largest area of remnant forest centred on Norfolk Island National Park (Director of National Parks Citation2020). A variety of introduced species are present on Norfolk Island, including Polynesian Rats (introduced ~800 years ago) and Black Rats (introduced in the 1940s) (Robinson Citation1978; Anderson and White Citation2001). Polynesian Rats and Black Rats can be abundant across the island, while House Mice Mus musculus tend to be more abundant within the human-modified landscape (Nance et al. Citation2023). All rodent species on Norfolk Island are of a similar size to their counterparts that occupy continental systems (M Wilson, pers obs.) Despite the relatively small size of the National Park (~460 ha), the intact forest communities support a concentration of threatened species including almost all Norfolk Island Robins, Slender-billed White-eyes, breeding Green Parrots Cyanoramphus cookii and Norfolk Island Moreporks (Sperring et al. Citation2021; Gautschi et al. Citation2022; Nance et al. Citation2023).
Owl capture and pellet collection
Fieldwork was conducted from October 2020 to January 2021 (morepork breeding season) and May to June 2021 (non-breeding season). Moreporks were captured using mist-nets at dusk with broadcasts of morepork recordings as a lure. Individuals that exceeded a mass of 100 g were fitted with a Lotek Pinpoint VHF-75 tracker (total mass 3.5 g, 1.8–2.4% of total body mass consistent with ethical recommendations; Caccamise and Hedin Citation1985). Trackers were attached to the dorsal surface of the two central tail feathers with Tesa® tape and pre-programed to emit a VHF beacon during daylight hours. For individual owls that were not recaptured for removal of the tracker, the trackers would have been shed during the next tail moult.
Daytime roosts for each morepork were located using a handheld Yagi antenna and Lotek Biotracker receiver unit over a period of 2–8 weeks. Cotton sheets of a neutral colour (~1.5 m × 1.5 m) were affixed below identified roost sites, 1–2 m off the ground. Sheets were checked every one to five days and all pellets (Image S1) and scats were collected. Samples were stored in 70% ethanol in sterile vials and held at room temperature.
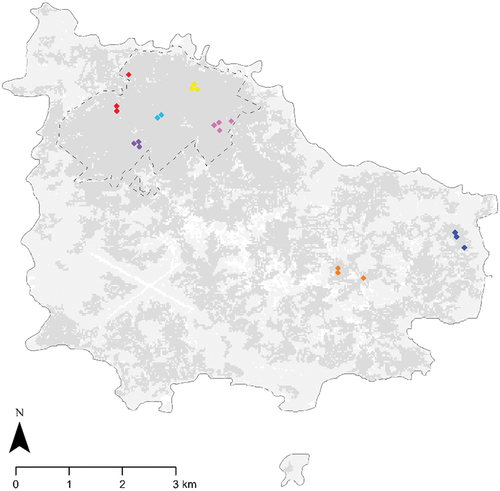
Moreporks were considered to be in a pair if two individuals were using the same roost during the tracking period. Where a pellet was collected in these settings, it was not possible to determine from which individual the sample originated, and thus paired owls were treated as one sample source. Sample locations were classified as ‘National Park’ or ‘modified landscape’, depending on the location of their roosts and territory.
Visual analysis
All pellets and a small reference collection of arthropods were examined using sterile single-use petri dishes under a dissecting microscope in the laboratory (Image S2). Invertebrate prey remains were first identified to order, and vertebrate remains were identified to class where possible. Unique taxa from each invertebrate order were either identified to species level or recorded with a unique identifier. Identification was aided by the reference collection and the assistance of experts from relevant taxonomic fields. The abundance of each prey was assessed by counting the minimum possible number of individuals represented by identifiable remains.
eDNA analysis
DNA was extracted from scats, and pellets collected from May to June 2021 using the Qiagen PowerSoil Pro DNA extraction kit (Qiagen, Clayton, Australia).
Metabarcoding
Universal vertebrate and invertebrate assays targeting part of the 12S mitochondrial DNA gene region were used to characterise vertebrates and invertebrates in the scat and pellet samples. We used a two-step PCR library construction method (see McColl‐Gausden et al. Citation2021). For vertebrate analysis, the first round of PCR employed gene-specific primers (vertebrate 12S; Riaz et al. Citation2011) to amplify the target region. For invertebrate analysis, the first round of PCR employed gene-specific COI (mitochondrial) primers (Zeale et al. Citation2011). The second round incorporated sequencing adapters and unique barcodes for each sample–amplicon combination included in the library. Negative control samples were also included during library construction. Negative controls consisted of the extraction negative as well as PCR negatives, where nuclease-free water was used in place of DNA during both rounds of PCR. Sequencing was carried out on an Illumina iSeq 100 machine using the iSeq i1 reagent kit v2 (Illumina, San Diego, CA, USA) with paired-end reads.
Vertebrate bioinformatic analysis
Following quality control filtering to remove primer sequences, truncated reads and low-frequency reads, DNA sequences were deduplicated, and all unique sequences were retained and assigned a running Operational Taxonomic Unit (OTU) number. Taxonomic assignment was performed with VSEARCH software (Rognes et al. Citation2016), whereby each OTU cluster was assigned a species identity using a threshold of 95% by comparing against a reference sequence database. Where a species could not be assigned (i.e. the reference database was deficient and/or taxa were poorly characterised), taxonomic assignments were manually vetted by first obtaining a list of possible species through BLASTN searches against the public repository GenBank (www.ncbi.nlm.nih.gov). Species were then discounted on the basis of their geographic distribution using information from the Atlas of Living Australia (ALA). In cases where an OTU could not be adequately resolved to a single species (due to shared haplotypes for instance), it was assigned to a higher taxonomic rank. Simultaneously, in cases where there was the possibility of ‘over-assignment’ (assignments to species with little data corroborating their occurrence in Australia), we also assigned such OTUs to a higher taxonomic level. Examples of such over-assignments can occur when congeners (or confamilials) are present in Australia but there is little to no genetic sequencing available for individual Australian species.
Invertebrate bioinformatic analysis
Amplicon pools were first demultiplexed based on the unique barcodes that identified individual samples. Reads R1 and R2 from the paired-end sequencing were merged using the fastq-mergepairs function in VSEARCH (Edgar Citation2016), retaining only merged reads flanked by matches to the gene-specific COI primers. Following quality control filtering to remove primer sequences, truncated reads and low-frequency reads, DNA sequences were deduplicated, and all unique sequences were retained and assigned a running OTU number. Taxonomic assignment was performed with VSEARCH’s SINTAX algorithm (Edgar Citation2016). Each OTU was assigned a taxonomic identity using a threshold of 80% by comparing against the MIDORI2 reference sequence database (Leray et al. Citation2022), supplemented by BLAST searches. In cases where OTUs could not be adequately assigned to a species, they are assigned to the lowest taxonomic rank possible.
Data filtering
Low-level detections that were likely to represent contamination events from a natural source were excluded by removing detections with fewer than 50 sequencing reads (Dully et al. Citation2021). Taxa detected with an adult body length <10 mm were also excluded (as possible secondary detections) as moreporks are expected to mostly consume prey larger than this threshold (Denny Citation2009).
Data analysis
We compared detections from both analysis methods (eDNA metabarcoding and visual analysis of the same pellets) and sample types (eDNA analysis of scats and pellets) to determine prey taxa richness per owl. Specifically, we investigated taxon richness within avifauna, rodents, and various invertebrate orders. The difference in richness (∆ richness) between analysis methods, and sample types was calculated per owl for each taxonomic group. The minimum, maximum and average ∆ richness was then determined per taxonomic group.
Using visual analysis outputs, Fisher’s exact tests were run to compare the proportion of rodents detected in pellets between seasons and habitat. We also ran a Fisher’s exact test to compare the proportion of vertebrate and invertebrate detections between eDNA sample types. All analyses were implemented in R, and significance was tested at the 0.05 level (R Core Team Citation2023).
Results
Samples were collected from seven Norfolk Island Moreporks, with each bird utilising two or three day-roosts (). Two owls occurred as a pair and two owls occupied territories entirely outside of the National Park. With a population estimate of 25 individuals, samples are representative of approximately one-third of the population (F. Sperring unpubl. data). A total of 24 pellets (2–7 per owl) and 19 scats (2–6 per owl) were collected in autumn from five owls, and 89 pellets (5–25 per owl) were collected in spring from six owls.
Figure 1. Roost sites for each Norfolk Island Morepork sampled across Norfolk Island during the austral spring of 2020 and austral autumn of 2021. The grey shaded areas represent vegetation. The National Park is outlined with a dashed line. Each colour represents a different owl. The owls marked in pink and purple were sampled only in spring. The owl marked in light blue was sampled only in autumn.
Vertebrate consumption
Using eDNA metabarcoding, prey taxa richness for each owl was between two and five vertebrate taxa (). All individuals were found to have consumed rodents. Rodents were also detected in 44% of all samples, with a more refined identification to Rattus sp. detected in 32% of samples. House Mice were consumed by two owls. The proportion of pellets that contained rodents did not differ between season or habitat (χ2(1, 113) = 0.016, p = 0.90 and χ2(1, 113) < 0.001, p = 1, respectively; visually screened pellets only).
Table 1. Dietary items consumed by Norfolk Island Moreporks as shown by eDNA analysis. Numbers represent the total number of pellets or scats that contained a particular prey item. Scat samples are shown in bold. Detections from scat and pellet samples collected on the same day are included. Total count represents the total number of detections of each species, with samples from the same or consecutive day excluded. Paired owls are identified with ‘a/b’.
The eDNA metabarcoding analysis detected Psittacidae sp. (either the introduced Crimson Rosella Platycercus elegans or endemic and threatened Green Parrot) in the pellets of one owl and Zosterops sp. (either the Silvereye Z. lateralis or the Vulnerable Slender-billed White-eye) was detected as prey for four of five owls, and in 28% of all samples screened. Visual analysis was unable to identify bird taxa below the avian order that had been consumed as prey. The contents of a nest box that supported a breeding pair of moreporks in 2019 included bird bands (issued by the Australian Bird and Bat Banding Scheme) that had been placed on a Slender-billed White-eye and a Norfolk Robin, indicating both species had been brought to the nest box as prey during that period.
Invertebrate consumption
Using all samples screened with metabarcoding, invertebrate prey richness was between three and seven taxa per owl (). Araneae (spiders) were consumed by all owls. All owls also consumed at least two of seven Lepidoptera taxa (moths and butterflies) taken as prey. Stylommatophora (land snails and slugs) were detected in three samples from one owl occupying territory entirely within the National Park. Visual analysis of pellets identified Coleoptera (beetles), Orthoptera (grasshoppers and crickets) and Lepidoptera in the diet of moreporks (). Coleoptera were found in 75% of pellets in autumn and 84% of pellets in spring while Orthoptera were found in 92% of pellets in autumn and 62% in spring.
Table 2. Abundance of each prey taxa identified for individual Norfolk Island Morepork using visual analysis of pellets in the breeding and non-breeding seasons. Abundance indicates the minimum possible number of individuals of each prey taxa consumed per owl. Paired owls are identified with ‘a/b’. The total number of pellets collected per individual/pair is shown.
Comparison of methods
Metabarcoding of pellets and scats identified 25 taxa in the diet of moreporks in the single season that was screened using this technique. By contrast, visual screening of pellets identified just nine taxa across combined breeding and non-breeding seasons. Where the same pellet was screened using both methods, each vertebrate taxon that was identified visually was also detected using eDNA metabarcoding. Metabarcoding detected more species of bird that were consumed as prey and visual analysis detected more species/taxa of Orthoptera and Coleoptera that were consumed as prey ().
Figure 2. Average ∆ richness for each taxonomic group comparing (a) visual and eDNA analysis and (b) eDNA analysis of scats and pellets collected from Norfolk Island Moreporks. In Figure 1(a), negative values represent greater taxa richness detected with visual analysis and positive values represent greater richness with eDNA. In Figure 1(b), negative values represent greater taxa richness identified in scats and positive values represent greater richness in pellets. Minimum and maximum ∆ richness per owl are shown with horizontal bars.
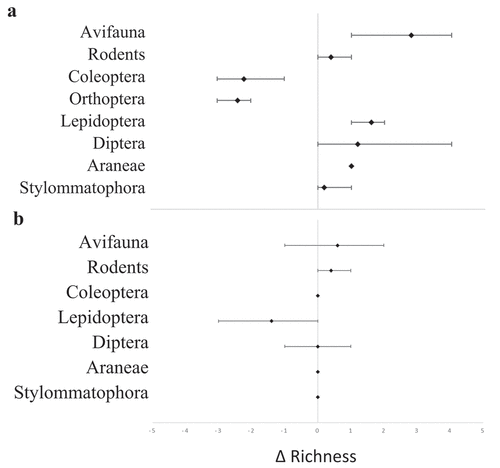
Richness for each taxonomic group and total taxa richness was similar for analyses of pellets and scats (pellets = 19 taxa (n = 15), scats = 17 (n = 19)) (). At a finer taxonomic resolution, the screening of pellets detected significantly more vertebrate taxa than scats, while the screening of scats detected significantly more invertebrate taxa than pellets (p = 0.048) (vertebrate richness: scats = 4, pellets = 11. Invertebrate richness: scats = 13, pellets = 8).
Discussion
In this study, we quantified the prey frequency and dietary breadth of the critically endangered Norfolk Island Morepork. Molecular screening of pellets and scats was an effective method for determining the diet of the owls and, in combination with visual analysis, revealed that all moreporks consumed invasive rodents. In the presence of an ongoing rodent control programme, these results highlight a conundrum for conservation managers at this site. The control of invasive rodents is considered essential for the recovery of threatened species on Norfolk Island, yet this same management programme poses a genuine threat through secondary poisoning to the tiny remaining morepork population.
The rodent control conundrum
Almost half of all dietary samples collected during the non-breeding period and screened using metabarcoding contained rodents (44%), and there was no significant difference in the proportion of rodent-positive samples between seasons or habitat using visual analysis. In other species, consumption of rodents has been posited to be largely influenced by two factors: resource requirements for breeding, and prey abundance (Olsen Citation2012; McDonald and Pavey Citation2014; Olsen et al. Citation2023, Citation2024). Invertebrate abundance is often lower in winter, which is thought to cause an increase in the uptake of rodent consumption (Stephenson Citation1998; Trost et al. Citation2008; Olsen et al. Citation2023). However, during spring, the elevated energetic requirements associated with breeding can also lead to an increase in the consumption of vertebrates (Olsen Citation2012; Olsen et al. Citation2023). This is supported by the absence of House Mice in the diet of Norfolk Island Moreporks occupying habitat within the National Park. While House Mice are an appropriate prey size, they are found in extremely low densities within the National Park (Nance et al. Citation2023). We suggest the consistent prevalence of rodents in the diet of the Norfolk Island Morepork may best be explained by the documented high abundance and year-round and island-wide availability of rodents (Nance et al. Citation2023).
Given the frequent consumption of rodents by the owls, and the use of second-generation rodenticides across Norfolk Island, secondary poisoning can be inferred to pose a genuine risk to the Norfolk Island Morepork. There is a growing body of research suggesting that secondary impacts of rodenticides are greater than previously thought (Lohr and Davis Citation2018), and elsewhere the incidence of secondary poisoning for boobooks, moreporks and larger Ninox species that consume rodents is well documented (Stephenson et al. Citation1999; Lohr Citation2018; Cooke et al. Citation2023). Locally, the only known Norfolk Island Morepork chick that hatched between 2011 and 2019 died in a nest box due to suspected secondary poisoning, when second-generation baits were used extensively within the National Park. More recently, an adult with symptoms of secondary poisoning (possibly Alphachloralose poisoning associated with feral chicken control; F. Sperring unpubl. data) was rehabilitated in 2021. Given moreporks and most other threatened fauna species occur in sympatry within the National Park, it is not possible to implement a spatially explicit rodent control programme that targets areas of high conservation priority for rodent management and avoids moreporks. Further exploration of owl-safe rodent control methods should be considered a priority action.
While Norfolk Island Moreporks will likely benefit from a reduction in the use of second-generation rodenticides, these same toxins are currently considered essential to suppress rodent populations and reduce nest predation of threatened songbirds (Nance et al. Citation2023). Invasive rodents on Norfolk Island are implicated in the extinction of many bird species and are responsible for the majority of nest failures in the endemic songbirds that persist (Nance et al. Citation2021, Citation2023). Nest survival of threatened songbirds is higher in areas with more intensive baiting, emphasising the importance of the current control methods (Nance et al. Citation2023). While rodent control methods are continually being improved, there are currently no methods other than second-generation baits that effectively suppress rodents across large areas (Howald et al. Citation2007; Gronwald and Russell Citation2022; DIISE Citation2023). Thus, the dependence of Norfolk Island songbirds on a programme that utilises second-generation toxins may continue for some time.
Given the breadth of the threat that invasive rodents pose to biota on Norfolk Island, their management must remain an ongoing priority. Therefore, effective rodent control strategies that simultaneously minimise the risk to moreporks warrant urgent exploration. Firstly, a non-toxic control measure currently being trialled on Norfolk Island is the use of self-resetting kill traps (e.g. Goodnature A24 or AT220 traps). While this technique is not uniformly successful, these traps have had some success in areas with large rodent populations (Peters et al. Citation2014; Gronwald and Russell Citation2022). Secondly, olfactory misinformation may also be used to manipulate rodent behaviour to disregard certain cues. For example, rodents can be manipulated to no longer associate the smell of songbird nests with a reward, consequently avoiding such cues in the future and minimising the predation risk to songbirds (Price and Banks Citation2012). Finally, a promising toxicant under investigation is cholecalciferol (vitamin D3), which raises blood calcium levels causing death through heart failure (Hix et al. Citation2012). This toxin has proven to be effective for rodent control and shows promising results for a reduced risk of secondary poisoning (Eason et al. Citation2000; Noh et al. Citation2023). Each of these approaches has the potential to reduce the impact of rodents, while minimising the risk to non-target species. However, further research and development is required. In the interim, population modelling of threatened songbirds and a formal risk assessment for Norfolk Island Moreporks under different baiting scenarios would be valuable to inform the optimal baiting strategy for threatened species management.
There are also some more ambitious strategies that may be worthy of consideration, notwithstanding considerable logistical, financial and social challenges. These include the development of a predator-free enclosure encompassing the National Park and surrounding forested areas, or an island-wide rodent eradication (Howald et al. Citation2007). For any rodent eradication attempt, temporary captive care of moreporks may present as a viable strategy (O’Dwyer et al. Citation2023). Future techniques such as gene drive technology may also provide a more species-specific, toxin-free eradication option (Leitschuh et al. Citation2017; Prowse et al. Citation2017). However, projected timelines to success might extend across multiple decades and consequently render this approach inviable. Finally, more intensive restoration efforts on nearby Phillip Island, which is rodent free but largely devoid of forested areas, may present an alternative pathway, or a complementary strategy, to increase the resilience of both morepork and songbird populations (Commonwealth of Australia Citation2024).
Norfolk Island exemplifies the importance of an integrated, ecosystem-wide approach for threatened species management. We provide evidence that the Norfolk Island Morepork consumes four of five remaining endemic songbirds (two of which are threatened), and possibly the Endangered Green Parrot. In a scenario where rodent abundance has been substantially reduced, a dietary shift of Norfolk Island Moreporks to other species (whose abundance may increase in the abundance of rodents) is likely (Denny Citation2009). The effective management of anthropogenic threats to endemic birds, namely of invasive mammals and habitat loss, should ensure that these same populations are robust to natural predation by moreporks (Salo et al. Citation2007; Gautschi et al. Citation2022). Additional goals for ecological restoration might include the return of nocturnally active reptile species and a species of giant centipede from Phillip Island to Norfolk Island. The reptile species, and probably the giant centipede, appear to have been extirpated from Norfolk Island because of predation by invasive rodents, and were likely to have featured prominently in the diet of the Norfolk Island Morepork prior to human settlement (Olsen Citation2012; Director of National Parks Citation2020).
Methods comparison
Comparing eDNA analysis with visual inspection, total taxon richness was similar between pellets and scats, scat analysis detected significantly more invertebrate taxa and pellet analysis detected more vertebrate taxa. Feathers, bones and fur are indigestible and must be regurgitated (Smith and Richmond Citation1972), whereas soft bodied invertebrates are likely to pass through the digestive system (Hill and Lill Citation1998). Consequently, taxon-specific digestive processes may influence the residual eDNA in different sample types.
Relative to visual analysis, eDNA identified three times as many prey taxa, with particular improvements to the identification of soft-bodied prey. For example, Stylommatophora, Diptera and Araneae were detected only through metabarcoding. Stylommatophora (land snails and slugs), are particularly interesting as putative prey of the Norfolk Island Morepork as these have not previously been identified in the diet of moreporks or boobooks. Our results are consistent with previous studies highlighting the value of eDNA analysis for a more comprehensive and refined assessment of dietary composition and breadth (Sousa et al. Citation2019; Hoenig et al. Citation2022).
Visual analysis identified more Coleoptera and Orthoptera taxa than metabarcoding. This may be explained by the incompleteness of the reference sequence database (Beng and Corlett Citation2020) which did not contain many of the invertebrate species’ endemic to Norfolk Island. Since reference sequences are often available only for a few genes for most species, targeted marker regions cannot always accurately resolve all groups, even to higher taxonomic levels (Liu et al. Citation2017; Beng and Corlett Citation2020). A more complete sequence database would likely improve the identification of these orders. We highlight the value of developing eDNA libraries for island ecosystems to investigate trophic interactions. In settings where trophic relationships may directly inform management considerations, we highlight the value of applying multiple screening strategies to investigate species diet.
Conclusion
During both the breeding and non-breeding season, Norfolk Island Moreporks consumed invasive rodents that were subject to control measures using toxic baits. This places a critically endangered owl at risk of secondary poisoning, with anecdotal evidence that this is indeed occurring. The conundrum for managers is that the current rodent baiting programme on Norfolk Island is known to improve the nesting success of endemic songbirds. This management challenge therefore requires a considered response to prevent the loss of songbirds, while minimising the threat of secondary poisoning to moreporks. We recommend managers urgently explore novel approaches and innovative technologies to control rodents effectively and safely. Our study highlights the interconnectedness of systems through food webs and that a targeted understanding of trophic interactions can be essential to inform effective whole-of-system conservation actions.
Supplemental Material 2
Download MS Word (1.1 MB)Supplemental Material 1
Download MS Word (1.1 MB)Acknowledgments
We pay our respect to the Norf’k Ailen Kaunsl’ Eldas and extend our recognition to all Pitkern descendants who call Norf’k home. We acknowledge the contribution of Parks Australia and Norfolk Island National Park, Norfolk Island Regional Council and the Norfolk Island community for their cooperation and assistance during this research. Particular thanks go to Nigel Greenup, Margaret Christian, Mark Hallam and Jolene Oliver. Rohan Bilney and the CSIRO Insect Collections team provided invaluable assistance with visual taxa identification. We thank Anthony van Rooyen for eDNA metabarcoding. Penny Olsen, Allie Nance, Rebecca Pirzl and three anonymous referees provided valuable feedback on the manuscript. This research was funded through the National Environmental Science Program and by Director of National Parks. All research was approved by Parks Australia and conducted under Monash University Animal Ethics approval 2021-25691. FS was supported by an Australian Government Research Training Program Scholarship. The authors report there are no competing interests to declare.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed at https://doi.org/10.1080/01584197.2024.2335397.
Additional information
Funding
References
- Anderson, A., and White, P. (2001). Approaching the prehistory of Norfolk Island. Records of the Australian Museum 27(53), 1–9. doi:10.3853/j.0812-7387.27.2001.1335
- Beng, K. C., and Corlett, R. T. (2020). Applications of environmental DNA (eDNA) in ecology and conservation: Opportunities, challenges and prospects. Biodiversity and Conservation 29(7), 2089–2121. doi:10.1007/s10531-020-01980-0
- Caccamise, D. F., and Hedin, R. S. (1985). An aerodynamic basis for selecting transmitter loads in birds. The Wilson Bulletin 97, 306–318.
- Canale, G. R., and Bernardo, C. S. S. (2016). Predator-prey interaction between two threatened species in a Brazilian hotspot. Biota Neotropica 16, e0059. doi:10.1590/1676-0611-bn-2015-0059
- Cavallo, C., Chiaradia, A., Deagle, B. E., McInnes, J. C., Sánchez, S., and Hays, G. C., et al. (2018). Molecular analysis of predator scats reveals role of salps in temperate inshore food webs. Frontiers in Marine Science 5, 381. doi:10.3389/fmars.2018.00381
- Commonwealth of Australia. (2024). ’Norfolk Island Region Threatened Species Recovery Plan.’ (Department of Climate Change, Engery, The Environment and Water: Canberra.)
- Cooke, R., Wallis, R., Hogan, F., White, J., and Webster, A. (2006). The diet of Powerful Owls (Ninox strenua) and prey availability in a continuum of habitats from disturbed urban fringe to protected forest environments in south-eastern Australia. Wildlife Research 33(3), 199–206. doi:10.1071/WR05058
- Cooke, R., Whiteley, P., Death, C., Weston, M. A., Carter, N., and Scammell, K., et al. (2023). Silent killers? The widespread exposure of predatory nocturnal birds to anticoagulant rodenticides. Science of the Total Environment 904, 166293. doi:10.1016/j.scitotenv.2023.166293
- Croll, D. A., Newton, K. M., McKown, M., Holmes, N., Williams, J. C., and Young, H. S., et al. (2016). Passive recovery of an island bird community after rodent eradication. Biological Invasions 18(3), 703–715. doi:10.1007/s10530-015-1042-9
- Denny, K. M. (2009). The diet of moreporks (Ninox novaeseelandiae) in relation to prey availability, and their roost site characteristics and breeding success on Ponui Island, Hauraki Gulf, New Zealand. Masters Thesis, Massey University, New Zealand.
- DIISE. (2023). The database of island invasive species eradications. Available at http://diise.islandconservation.org/. [Accessed July 2023].
- Director of National Parks. (2020). ’Norfolk Island National Park and Norfolk Island Botanical Garden Management Plan 2020.’ (Director of National Parks: Canberra.)
- Doherty, T. S., Glen, A. S., Nimmo, D. G., Ritchie, E. G., and Dickman, C. R. (2016). Invasive predators and global biodiversity loss. Proceedings of the National Academy of Sciences 113, 11261–11265. doi:10.1073/pnas.1602480113
- Driver, E. C. (1949). Mammal remains in owl pellets. The American Midland Naturalist 41(1), 139–142. doi:10.2307/2422021
- Dully, V., Wilding, T. A., Mühlhaus, T., and Stoeck, T. (2021). Identifying the minimum amplicon sequence depth to adequately predict classes in eDNA-based marine biomonitoring using supervised machine learning. Computational and Structural Biotechnology Journal 19, 2256–2268. doi:10.1016/j.csbj.2021.04.005
- Eason, C. T., Wickstrom, M., Henderson, R., Milne, L., and Arthur, D. (2000). Nontarget and secondary poisoning risks associated with cholecalciferol. New Zealand Plant Protection 53, 299–304. doi:10.30843/nzpp.2000.53.3699
- Edgar, R. C. (2016). SINTAX: A simple non-Bayesian taxonomy classifier for 16S and ITS sequences. bioRxiv, 074161. doi:10.1101/074161
- Erickson, W., and Urban, D. (2004). ’Potential Risks of Nine Rodenticides to Birds and Nontarget Mammals: A Comparative Approach.’ (US Environmental Protection Agency, Office of Prevention, Pesticides and Toxic Substances: Washington.)
- Fisher, P., Campbell, K. J., Howald, G. R., and Warburton, B. (2019). Anticoagulant rodenticides, islands and animal welfare accountancy. Animals 9, 919. doi:10.3390/ani9110919
- Gautschi, D., Heinsohn, R., Crates, R., Macgregor, N. A., Wilson, M., and Stojanovic, D. (2022). Utilization of modified and artificial nests by endemic and introduced parrots on Norfolk Island. Restoration Ecology 30(5), e13586. doi:10.1111/rec.13586
- Gronwald, M., and Russell, J. (2022). Behaviour of invasive ship rats, Rattus rattus, around goodnature A24 self-resetting traps. Management of Biological Invasions 13(3), 479–493. doi:10.3391/mbi.2022.13.3.02
- Hadler, M. R., and Buckle, A. P. (1992). Forty-five years of anticoagulant rodenticides–past, present and future trends. Paper presented at the Proceedings of the Vertebrate Pest Conference, California.
- Hill, F. R., and Lill, A. (1998). Diet and roost site characteristics of the Christmas Island Hawk-Owl Ninox natalis. Emu 98, 227–233. doi:10.1071/MU98031
- Hix, S., Aylett, P., Shapiro, L., MacMorran, D., Eason, C., and Sam, S., et al. (2012). Low-dose cholecalciferol bait for possum and rodent control. New Zealand Journal of Agricultural Research 55(3), 207–215. doi:10.1080/00288233.2012.665806
- Hoenig, B. D., Snider, A. M., Forsman, A. M., Hobson, K. A., Latta, S. C., and Miller, E. T., et al. (2022). Current methods and future directions in avian diet analysis. The Auk 139(1), 1–28. doi:10.1093/ornithology/ukab077
- Howald, G., Donlan, C. J., Galvan, J. P., Russell, J. C., Parkes, J., and Samaniego, A., et al. (2007). Invasive rodent eradication on islands. Conservation Biology 21(5), 1258–1268. doi:10.1111/j.1523-1739.2007.00755.x
- Huckle, K. R., Hutson, D. H., and Warburton, P. A. (1988). Elimination and accumulation of the rodenticide flocoumafen in rats following repeated oral administration. Xenobiotica 18(12), 1465–1479. doi:10.3109/00498258809042269
- Jones, H. P., Holmes, N. D., Butchart, S. H., Tershy, B. R., Kappes, P. J., and Corkery, I., et al. (2016). Invasive mammal eradication on islands results in substantial conservation gains. Proceedings of the National Academy of Sciences 113, 4033–4038. doi:10.1073/pnas.1521179113
- Keitt, B., Campbell, K., Saunders, A., Clout, M., Wang, Y., and Heinz, R., et al. (2011). The global islands invasive vertebrate eradication database: A tool to improve and facilitate restoration of island ecosystems. Paper presented at the Island invasives: eradication and management, IUCN, Gland, Switzerland.
- Leitschuh, C. M., Kanavy, D., Backus, G. A., Valdez, R. X., Serr, M., and Pitts, E. A., et al. (2017). Developing gene drive technologies to eradicate invasive rodents from islands. Journal of Responsible Innovation 5, S121–S138. doi:10.1080/23299460.2017.1365232
- Leray, M., Knowlton, N., and Machida, R. J. (2022). MIDORI2: A collection of quality controlled, preformatted, and regularly updated reference databases for taxonomic assignment of eukaryotic mitochondrial sequences. Environmental DNA 4(4), 894–907. doi:10.1002/edn3.303
- Liu, S., Yang, C., Zhou, C., and Zhou, X. (2017). Filling reference gaps via assembling DNA barcodes using high-throughput sequencing—moving toward barcoding the world. GigaScience 6(12), gix104. doi:10.1093/gigascience/gix104
- Lohr, M. T. (2018). Anticoagulant rodenticide exposure in an Australian predatory bird increases with proximity to developed habitat. Science of the Total Environment 643, 134–144. doi:10.1016/j.scitotenv.2018.06.207
- Lohr, M. T., and Davis, R. A. (2018). Anticoagulant rodenticide use, non-target impacts and regulation: A case study from Australia. Science of the Total Environment 634, 1372–1384. doi:10.1016/j.scitotenv.2018.04.069
- Marquez, A., Khalil, R. A., Fourel, I., Ovarbury, T., Pinot, A., and Rosine, A., et al. (2019). Resistance to anticoagulant rodenticides in Martinique could lead to inefficient rodent control in a context of endemic leptospirosis. Scientific Reports 9(1), 13491. doi:10.1038/s41598-019-49661-5
- Maser, C., and Brodie, E. D. (1966). A study of owl pellet contents from Linn, Benton and Polk counties, Oregon. The Murrelet 47(1), 9–14. doi:10.2307/3536232
- McColl‐Gausden, E. F., Weeks, A. R., Coleman, R. A., Robinson, K. L., Song, S., and Raadik, T. A., et al. (2021). Multispecies models reveal that eDNA metabarcoding is more sensitive than backpack electrofishing for conducting fish surveys in freshwater streams. Molecular Ecology 30(13), 3111–3126. doi:10.1111/mec.15644
- McDonald, P., and Pavey, C. (2014). Exploiting boom times. Southern boobook owl Ninox novaeseelandiae diet during a rodent irruption in central Australia. Australian Zoologist 37(2), 234–237. doi:10.7882/AZ.2014.024
- Menning, D. M., Uher-Koch, B. D., Flamme, M. J., Simmons, T., Schmutz, J. A., and Talbot, S. L. (2023). eDNA metabarcoding analyses of diet in Yellow-billed Loons of Northern Alaska. Colonial Waterbirds 45(2), 159–166. doi:10.1675/063.045.0206
- Nance, A., Mitchell, W., Clarke, R., Wilson, M., Brown, S., and Macgregor, N., et al. (2021). Norfolk Island Robin (Petroica multicolor). In ’The Action Plan for Australian Birds 2020.’ (Eds S. Garnett and G. Baker) pp. 741–744. (CSIRO Publishing: Melbourne.) doi:10.1071/9781486311910
- Nance, A., Wilson, M., Cook, C., and Clarke, R. H. (2023). Arboreal activity of invasive rodents: Conservation implications for the control of an island pest. Pacific Conservation Biology 30, C23011. doi:10.1071/PC23011
- Nance, A. H., Mitchell, W. F., Dawlings, F., Cook, C., and Clarke, R. (2023). Rodent predation and specialised avian habitat requirements drive extinction risk for endemic island songbirds in the south-west Pacific. Emu-Austral Ornithology 123, 1–15. doi:10.1080/01584197.2023.2228350
- Noh, A. A. M., Ahmad, A. H., and Salim, H. (2023). Efficacy of cholecalciferol rodenticide to control Wood Rat, Rattus tiomanicus and its secondary poisoning impact towards Barn Owl. Tyto javanica javanica Scientific Reports 13, 2854. doi:10.1038/s41598-023-29499-8
- O’Dwyer, T. W., Carlile, N., O’Neill, L., Fairlamb, H., and Bower, H. (2023). Protection and mortality of non-target terrestrial bird species during the eradication of rodents on Lord Howe Island. Biological Invasions 26, 151–167. doi:10.1007/s10530-023-03161-w
- Olsen, J. (2012). ’Australian High Country Owls.’ (CSIRO Publishing: Melbourne.)
- Olsen, J., Trost, S., Rose, A. B., Fuentes, E. E., and Debus, S. (2023). Diet of Southern Boobooks Ninox boobook and its relationship with breeding in Canberra, Australian capital territory, 1993–2019. Corella 47, 45–55.
- Olsen, J., Trost, S., Rose, A. B., Fuentes, E. E., and Debus, S. (2024). Influence of hunting grounds on prey taken in adjacent territories of breeding southern boobooks Ninox boobook in the Australian capital territory. Corella 48, 28–31.
- Olsen, P. (1997). ’Recovery Plan for the Norfolk Island Boobook Owl, Ninox Novaeseelandiae Undulata.’ (Australian National University: Canberra.)
- Olsen, P. D. (1996). Re-establishment of an endangered subspecies: The Norfolk Island Boobook Owl Ninox novaeseelandiae undulata. Bird Conservation International 6(1), 63–80. doi:10.1017/S0959270900001313
- Park, B., Scott, A., Wilson, A., Haynes, B., and Breckenridge, A. (1984). Plasma disposition of vitamin K1 in relation to anticoagulant poisoning. British Journal of Clinical Pharmacology 18(5), 655–662. doi:10.1111/j.1365-2125.1984.tb02526.x
- Peters, D. H., Schumacher, K., Schumacher, R. J., and Baigent, D. W. (2014). Goodnature automatic traps for vertebrate pest control: Field trials using new kill traps targeting animal pests in New Zealand. Proceedings of the Vertebrate Pest Conference 26, 404–410. doi:10.5070/V426110618
- Price, C. J., and Banks, P. B. (2012). Exploiting olfactory learning in alien rats to protect birds’ eggs. Proceedings of the National Academy of Sciences 109, 19304–19309. doi:10.1073/pnas.1210981109
- Prowse, T. A., Cassey, P., Ross, J. V., Pfitzner, C., Wittmann, T. A., and Thomas, P. (2017). Dodging silver bullets: Good CRISPR gene-drive design is critical for eradicating exotic vertebrates. Proceedings of the Royal Society B: Biological Sciences 284, 20170799. doi:10.1098/rspb.2017.0799
- Quasim, S., MacDonald, A. J., and Sarre, S. D. (2018). Towards more efficient large-scale DNA-based detection of terrestrial mammal predators from scats. Mammal Research 63(3), 387–393. doi:10.1007/s13364-018-0369-x
- R Core Team. (2023). ’R: A Language and Environment for Statistical Computing (4.2.2).’ (R Foundation for Statistical Computing.) https://www.r-project.org/
- Riaz, T., Shehzad, W., Viari, A., Pompanon, F., Taberlet, P., and Coissac, E. (2011). ecoPrimers: Inference of new DNA barcode markers from whole genome sequence analysis. Nucleic Acids Research 39(21), e145–e145. doi:10.1093/nar/gkr732
- Robinson, D. (1978). ’Ecology and Management of the Scarlet Robin, White-Breasted White-Eye and Long-Billed White-Eye on Norfolk Island.’ (Melbourne: Department of Zoology, Monash University.)
- Roemer, G. W., and Wayne, R. K. (2003). Conservation in conflict: The tale of two endangered species. Conservation Biology 17, 1251–1260. doi:10.1046/j.1523-1739.2003.02202.x
- Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahé, F. (2016). VSEARCH: A versatile open source tool for metagenomics. PeerJ 4, e2584. doi:10.7717/peerj.2584
- Russell, J. C., Meyer, J.-Y., Holmes, N. D., and Pagad, S. (2017). Invasive alien species on islands: Impacts, distribution, interactions and management. Environmental Conservation 44(4), 359–370. doi:10.1017/S0376892917000297
- Salo, P., Korpimäki, E., Banks, P. B., Nordström, M., and Dickman, C. R. (2007). Alien predators are more dangerous than native predators to prey populations. Proceedings of the Royal Society B: Biological Sciences 274, 1237–1243. doi:10.1098/rspb.2006.0444
- Smith, C. R., and Richmond, M. E. (1972). Factors influencing pellet egestion and gastric pH in the barn owl. The Wilson Bulletin 84, 179–186.
- Soulé, M. E., Estes, J. A., Berger, J., and Del Rio, C. M. (2003). Ecological effectiveness: Conservation goals for interactive species. Conservation Biology 17(5), 1238–1250. doi:10.1046/j.1523-1739.2003.01599.x
- Sousa, L. L., Silva, S. M., and Xavier, R. (2019). DNA metabarcoding in diet studies: Unveiling ecological aspects in aquatic and terrestrial ecosystems. Environmental DNA 1(3), 199–214. doi:10.1002/edn3.27
- Sperring, V., Brown, S., Macgregor, N., Olsen, P., Clarke, R., and Wilson, M., et al. (2021). Norfolk Island Morepork Ninox novaeseelandiae undulata In ’The Action Plan for Australian Birds 2020.’ (Eds S. Garnett and G. Baker.) pp. 360–363. (CSIRO Publishing: Melbourne.) doi:10.1071/9781486311910
- St Clair, J. J. (2011). The impacts of invasive rodents on island invertebrates. Biological Conservation 144, 68–81. doi:10.1016/j.biocon.2010.10.006
- Stephenson, B. M. (1998). The ecology and breeding biology of morepork, Ninox novaeseelandiae, and their risk from secondary poisoning, in New Zealand. Masters Thesis, Massey University, New Zealand.
- Stephenson, B. M., Minot, E. O., and Armstrong, D. P. (1999). Fate of moreporks (Ninox novaeseelandiae) during a pest control operation on Mokoia Island, Lake Rotorua, North Island, New Zealand. New Zealand Journal of Ecology 23, 233–240.
- Threatened Species Scientific Committee. (2016). ’Conservation Advice Ninox Novaeseelandiae Undulata.’ (Norfolk Island Boobook Owl. Department of the Environment: Canberra.)
- Trost, S., Olsen, J., Rose, A., and Debus, S. J. (2008). Winter diet of Southern Boobooks Ninox novaeseelandiae in Canberra 1997–2005. Corella 32, 66–70.
- Van den Brink, N. W., Elliott, J. E., Shore, R. F., and Rattner, B. A. (2018). ’Anticoagulant Rodenticides and Wildlife.’ (Springer: Switzerland.)
- Wheeler, R., Priddel, D., O’Dwyer, T., Carlile, N., Portelli, D., and Wilkinson, I. (2019). Evaluating the susceptibility of invasive black rats (Rattus rattus) and House Mice (Mus musculus) to brodifacoum as a prelude to rodent eradication on Lord Howe Island. Biological Invasions 21(3), 833–845. doi:10.1007/s10530-018-1863-4
- Whittaker, R. J., and Fernández-Palacios, J. M. (2007). ’Island Biogeography: Ecology, Evolution, and Conservation.’ (Oxford University Press: Oxford.)
- Zeale, M. R., Butlin, R. K., Barker, G. L., Lees, D. C., and Jones, G. (2011). Taxon‐specific PCR for DNA barcoding arthropod prey in bat faeces. Molecular Ecology Resources 11, 236–244. doi:10.1111/j.1755-0998.2010.02920.x

