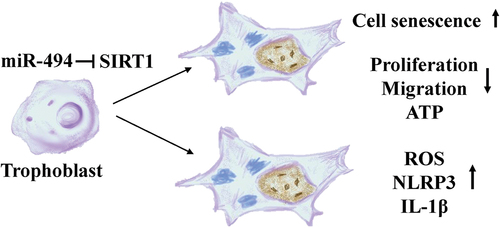
ABSTRACT
Objective
Although the mechanism underlying preeclampsia (PE) has been widely explored, the mechanisms related to senescence have not yet been fully revealed. Therefore, we investigated the role of the miR-494/longevity protein Sirtuin 1 (SIRT1) axis in PE.
Methods
Human placental tissue was obtained from severe preeclampsia (SPE) (n = 20) and gestational age-matched normotensive pregnancies (n = 20), and senescence-associated β-galactosidase (SAβG) and SIRT1 expression levels were measured. The TargetScan and miRDB databases predicted candidate miRNAs targeting SIRT1, and intersected with differentially expressed miRNAs in the GSE15789 dataset (p < 0.05, |log2FC|≥1.5). Subsequently, we showed that miRNA (miR)-494 expression was significantly elevated in SPE, revealing miR-494 as a candidate SIRT1-binding miRNA. A dual-luciferase assay confirmed the targeting relationship between miR-494 and SIRT1. The senescence phenotype, migration, cell viability, reactive oxygen species (ROS) production levels and inflammatory molecule expression levels were measured after miR-494 expression was altered. We conducted a rescue experiment using SIRT1 plasmids to further demonstrate the regulatory relationship.
Results
SIRT1 expression was lower(p < 0.01) and miR-494 expression was higher (p < 0.001) in SPE, and SaβG staining showed premature placental aging in SPE (p < 0.001). Dual-luciferase reporter assays revealed that miR-494 targeted SIRT1. Compared to control cells, HTR-8/SVneo cells with upregulation of miR-494 had remarkably downregulated SIRT1 expression (p < 0.001), more SAβG-positive cells (p < 0.001), cell cycle arrested (p < 0.05), and upregulated P21 and P16 expression (p < 0.01). miR-494 overexpression also decreased HTR-8/SVneo cell migration (p < 0.05) and ATP synthesis (p < 0.001), increased ROS levels (p < 0.001), and upregulated NLRP3 and IL-1β expression (p < 0.01). SIRT1-overexpressing plasmids partially reversed the effects of miR-494 overexpression in HTR-8/SVneo cells.
Conclusion
The miR-494/SIRT1 interaction plays a role in the mechanism of premature placental aging in PE patients.
Introduction
Preeclampsia (PE) is a syndrome unique to pregnancy that occurs in 2–8% of all pregnancies and is characterized by the new onset of hypertension and proteinuria in the latter half of gestation (Citation1). PE syndrome varies widely in its clinical phenotypic expression, but the final pathological change is diminished placental perfusion (Citation2,Citation3). Impaired placental blood flow results in a hypoxic environment and oxidative stress, triggering various forms of cellular stress, including cellular senescence (Citation4–8).
A recent bioinformatic analysis showed that an overall downregulation of antisenescence genes across preeclamptic placentas (Citation9), and premature placental aging might be involved in the pathogenesis of PE (Citation10,Citation11). Sirtuin 1 (SIRT1), an antisenescence gene (Citation12,Citation13), is dependent on intercellular nicotinamide adenosine dinucleotide (NAD) levels and is linked to placental development and epigenetic modifications (Citation14). SIRT1 may be a promising molecular biological target in placental aging. Regarding the epigenetics of aging, SIRT1 might affect placental development and trophoblast invasion through autophagy and senescence (Citation15). The downregulation of SIRT1 may accelerate placental senescence via targets contributing to the regulation of the cell cycle, extracellular matrix (ECM) production and cytoskeleton reorganization (Citation16). However, the specific mechanism of SIRT1 in placental aging in PE remains unclear.
MicroRNAs (miRNAs) are highly conserved endogenous small noncoding RNAs that play functional roles in gene expression at the posttranscriptional level by binding to the 3ʹ untranslated regions (3’-UTRs) of target mRNAs. The key role of miRNAs in aging has been highlighted in the past decade. miRNAs function as modulators of some pathways, including the NF-κB, mTOR, sirtuin, TGF-β and Wnt pathways, and play roles related to cellular senescence and age-related diseases (Citation17,Citation18). Many miRNAs are involved in placental development, and these miRNAs regulate the fate, function, differentiation and senescence of trophoblasts (Citation3,Citation19).
Therefore, our study aimed to investigate whether miRNAs targeting the antiaging gene SIRT1 contribute to premature placental senescence in PE to provide new insight into the mechanism of PE.
Materials and methods
Patient tissue collection
Placental samples were collected from women with severe PE (SPE) (n = 20) who underwent cesarean section at The First Affiliated Hospital of Guangxi Medical University from 2022 to 2023. Control placenta samples were obtained from women with normotensive pregnancies (n = 20) who underwent elective cesarean section. SPE was defined as severe hypertension (systolic blood pressure of 160 mmHg or diastolic blood pressure of 110 mmHg) or severe proteinuria (urinary protein excretion of 2 g per 24 hours). Women who had multifetal gestations, major fetal congenital anomalies, antepartum complications (diabetes mellitus, chronic hypertension, intrauterine infection), and histories of smoking and alcohol consumption were excluded. The clinical characteristics of the two groups of patients are compared in . This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Scientific Research Ethics Committee of The First Affiliated Hospital of Guangxi Medical University, Approval No. 2022-KT-011, and informed consent was obtained from all the participants.
Table 1. The demographics and clinical characteristicsa of patients in our study.
Cell culture
HTR-8/SVneo cells (Zhongqiao Xinzhou, China) were maintained in standard culture conditions at 37°C in a humidified 5% CO2 incubator. The culture medium (Gibco, USA) was RPMI 1640 supplemented with 10% heat-inactivated fetal bovine serum (FBS, Sijiqing, Zhejiang, China), 100 U/ml penicillin and 100 mg/ml streptomycin.
Cell transfection
Lipofectamine 3000 (Invitrogen, USA) was used for the transfection of these constructs into cells at 60–70% confluence based on the manufacturer’s instructions. After 48 h of transfection, the cells were used for follow-up experiments, and every experiment was repeated three times. The transfection concentrations were 50 nM for miR-494 and mimic negative control (NC), and the tissue culture plate diameter was 3.5 cm. For the rescue experiment, SIRT1 plasmids (1.17 μg), control (open reading frame) plasmids (1.17 μg), miR-494 (75 nM) and mimic NC (75 nM) were used. The miR-494 and miR mimic NC were synthesized by Sangon (Shanghai, China), and the plasmids were purchased from OBiO (Shanghai, China). All constructs were confirmed by DNA sequencing.
RT‑QPCR
Total RNA was extracted from tissues or cells with TRIzol (Invitrogen, USA) and reverse-transcribed (TaKaRa, Japan). The expression levels of miRNAs and SIRT mRNA were measured with SYBR Premix ExTaq with U6 and β-actin as reference controls, respectively. The primers are listed in .
Table 2. Primer sequences.
Western blot assay
Total protein was extracted from cells using RIPA buffer (Beyotime, China) supplemented with PMSF (Beyotime, China). The samples were separated by SDS‒PAGE, and the proteins were transferred to polyvinylidene difluoride (PVDF) membranes. The membranes were blocked with 5% nonfat milk (120 minutes), and immunoblotting was performed with antibodies against SIRT1 (0.8 μg/ml, Abmart, China), P21 (0.4 μg/ml, Abmart, China), P16 (0.4 μg/ml, Abmart, China), NLRP3 (0.8 μg/ml, WanleiBio, China), IL-1β (0.4 μg/ml, WanleiBio, China) and β-actin (1 μg/ml, Abmart, China). The protein bands were detected with ECL reagent (Abmart, China). With β-actin as the loading control, the relative expression of the target protein was calculated as the gray value ratio of the target protein to β-actin.
Immunohistochemistry and immunofluorescence microscopy
For immunohistochemistry, tissue sections were heated in citrate buffer in a microwave for antigen retrieval before exposure to primary antibodies. The tissue sections were incubated with a rabbit monoclonal anti-SIRT1 antibody (8 μg/ml, Abmart, China) at 4°C overnight. Biotinylated goat anti-rabbit IgG was used as the secondary antibody. For immunofluorescence staining, 24-well plates (2×103 cells/well) were incubated with a rabbit monoclonal anti-SIRT1 antibody (8 μg/ml Abmart, China) followed by a specific AF 488-conjugated goat anti-rabbit IgG secondary antibody (20 μg/ml, Abmart, China) for 1 h at room temperature and then counterstained with 4’-6-diamidino-2-phenylindole (DAPI, Solarbio, China) for 5 min. For the negative control, the primary antibody was replaced with an isotype control antibody. Images were captured using an Olympus B×53 microscope (Japan) and Evos FL Color Imaging System (US). A total of at least five different regions were used for each sample. Semiquantitative analysis of protein expression was performed by detecting the mean fluorescence intensity using ImageJ (Citation20,Citation21).
CCK-8 assay
Cells were seeded in 96-well plates (2×103 cells/well) and cultured under standard conditions for 72 h after transfection. Then, 110 µL/well of CCK-8 solution (Dojindo, Japan) (1:10) was added to the medium in each well. After an additional 2 h of incubation, the absorbance values of individual wells were assessed (450 nm) via a microplate reader.
Flow cytometry
After 48 h of transfection, cells were fixed (using 75% ethanol at −20°C), resuspended in phosphate-buffered saline (PBS) and stained with a Cell Cycle Detection Kit (MultiSciences, China). The cells were then analyzed by flow cytometry, and the results were assessed using Cell Quest Pro.
SAβG staining
After 48 h of transfection, HTR-8/SVneo cells were seeded in 6-well plates and stained with a Senescence β-Galactosidase Staining Kit (Beyotime, China). The images were captured by an Evos FL Color Imaging System (US) and NIS-Element (Japan). The ImageJ semiautomatic method was used to analyze the stained cells (Citation22).
Wound healing assay
Migration was assessed using a wound healing assay. Cells were seeded in 6-well plates (2×105 cells/well). After 48 h of transfection, a 100 µL pipette tip was used to scratch the cell monolayer. Fresh serum-free medium was added, and the cells were imaged with an inverted microscope (NIS-Element, Japan) at 0, 12 and 24 h to assess wound healing.
Detection of reactive oxygen species (ROS)
The intracellular levels of ROS were analyzed using the ROS-reactive fluorescent indicator 2,7-dichlorofluorescein diacetate (DCFH-DA, Beyotime, China). The cells were seeded in 6-well plates (1×105 cells/well) 48 h after transfection, treated with 10 µmol/L DCFH-DA for 20 min at 37°C in a 5% CO2 incubator, and washed with PBS. The samples were analyzed with an Evos FL Color Imaging System (US).
Firefly luciferase adenosine triphosphate (ATP) assay
ATP levels were quantified using the luciferin‑luciferase method. Cells were harvested after treatment, and ATP levels were measured using an ATP assay kit (Beyotime Institute of Biotechnology). In brief, the cells were lysed by adding 100 µL of ATP-releasing reagent. The lysates were incubated with the luciferin substrate and luciferase enzyme in the dark for 10 min to stabilize the luminescent signal. A fluorescence microplate reader was used to measure bioluminescence intensity.
Dual‑luciferase reporter gene assay
The wild-type (WT) SIRT1 3’-UTR sequence, a mutant (MUT) SIRT1 3’-UTR sequence with mutations in the putative miR-494 binding sites, and the Renilla sequence were synthesized by OBiO (Shanghai, China). Bioinformatics analyses were used to predict the binding sites of SIRT1 and miR-494. WT and MUT sequences of the 3’UTR of SIRT1 containing the miR-494 binding site were ligated into luciferase reporter vector constructs, and the MUT sequence was used to confirm the specificity of the interactions between SIRT1 and miR-494. HTR-8/SVneo cell cultures were cotransfected with WT-SIRT1 or MUT-SIRT1 and miR-494 or mimic NC. After cotransfection for 48 h, the binding activity was determined using a dual-luciferase reporter assessment kit (Promega, USA). The results are expressed as the ratio of the relative light units of firefly luciferase versus those of Renilla luciferase.
Statistical analysis
SPSS version 16.0 software (SPSS Inc, Chicago, IL, USA) and GraphPad Prism 7 were used for statistical analysis, and images were analyzed with ImageJ. For normally
distributed data, Student’s t test was used (for two-group comparisons); otherwise, the nonparametric Mann‒Whitney U test was employed. p < 0.05 was considered to indicate significance.
Results
SAβG and SIRT1 expression in normal and SPE placentas
SAβG activity is the most commonly used biomarker for cell senescence. Many more SAβG-positive cells in the syncytiotrophoblast and cytotrophoblast (arrowheads) were found in SPE placentas (, p < 0.001). SIRT1 is a longevity protein and was
Figure 1. SAβG and SIRT1 expression in normal (n = 20) and SPE (n = 20) placentas (×200). a. SAβG staining of human placenta sections. SAβG-positive cells in syncytiotrophoblasts and cytotrophoblasts (arrowheads) were increased in SPE placentas (p < 0.001). Scale bars, 50 µm. b. immunohistochemistry was used to measure SIRT1 protein expression in placental sections. SIRT1 was present in the syncytiotrophoblast and cytotrophoblast (arrowheads), but SIRT1 expression was lower in SPE placentas (p < 0.01). Scale bars, 50 µm. The data are presented as the mean ± SD, **p < 0.01, ***p < 0.001.
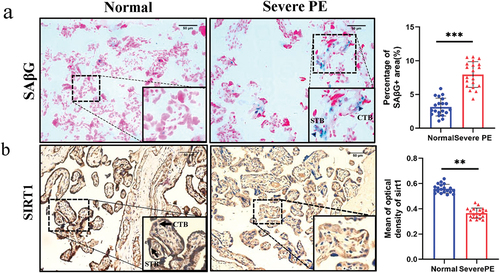
localized in the syncytiotrophoblast and cytotrophoblast (arrowheads). However, SIRT1 expression was lower in the placentas of SPE patients than in those of controls (, p < 0.01).
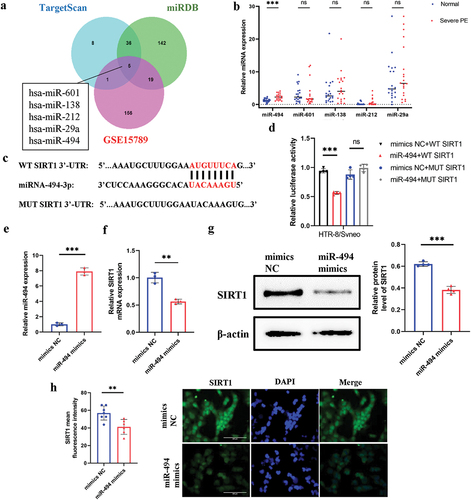
SIRT1 is the direct target of miR-494 in HTR-8/SVneo cells
We investigated the miRNAs that potentially mediate this process. Intersecting candidate miRNAs binding to SIRT1 were identified using two independent publicly available databases, namely, TargetScan (top 50) and miRDB, and differentially expressed miRNAs were identified in the GSE15789 dataset (p < 0.05, |log2FC|≥1.5). We obtained the following microRNAs: miR-601, miR-138, miR-212, miR-29a, and miR-494 (). The results showed that miR-494 expression was significantly elevated in SPE placentas (, p < 0.001). miR-494 binding sequences were identified in the 3”-UTR of SIRT1 mRNA (). Luciferase reporter assays revealed that miR-494 expression in HTR-8/SVneo cells suppressed the activity of the luciferase reporter fused to the WT 3’-UTR of SIRT1 but did not suppress that of a reporter fused to a mutant (MUT) version of the 3’-UTR (, p < 0.001). HTR-8/SVneo cells were transfected with miR-494 mimics for 48 h to upregulate the expression of miR-494 (, p < 0.001), and then we measured SIRT1 mRNA and protein levels. miR-494 significantly reduced SIRT1 mRNA (, p < 0.01) and protein (, p < 0.001) expression levels. Immunofluorescence staining showed the same results (, p < 0.01). These results suggest that SIRT1 is a direct target of miR-494 in HTR-8/SVneo cells.
Figure 2. SIRT1 is a direct target of miR-494 in HTR-8/SVneo cells. a. Prediction of miRnas that bind to SIRT1 (TargetScan top 50, miRDB, and GSE15789). b. RT‑qPCR analysis of miR-601, miR-138, miR-212, miR-29a, and miR-494 expression in placenta. c. The predicted binding sites of miR-494 in the 3’-UTR of SIRT1. d. Luciferase reporter assays. e. miR-494 expression after mimic transfection for 48 h. f. SIRT1 mRNA expression 48 h after miR-494 mimic transfection. g. SIRT1 protein expression after miR-494 mimic transfection for 48 h. h. Immunofluorescence staining of SIRT1 (green) after miR-494 mimic transfection for 48 h. Nuclei were stained with DAPI (blue). SIRT1 was present in the HTR-8/SVneo nucleus (×200). Scale bars, 200 µm. The data are presented as the mean ± SD from three independent experiments, and every experiment was repeated three times, *p < 0.05, **p < 0.01, ***p < 0.001.
MiR-494 induces a senescence phenotype in HTR-8/SVneo cells
After transfection with miR-494 mimics for 48 h, the number of SAβG-positive cells was increased over 5-fold (, p < 0.001). The G0/G1 phase fraction was increased (p < 0.05), and the S phase fraction decreased concomitantly in the miR-494 group ( p < 0.05), indicating cell cycle arrest. CCK-8 assays showed that cell proliferation was decreased after transfection with miR-494 mimics for 48 h and 72 h (, all p < 0.01). Western blotting (WB) showed that the protein expression of P21 and P16 was upregulated (, all p < 0.01). These findings suggest that miR-494 induces a senescence phenotype in HTR-8/SVneo cells and that this effect may be mediated by inhibition of SIRT1.
Figure 3. MiR-494 induces a senescent phenotype in HTR-8/SVneo cells a. miR-494 mimics increased the activity of SAβG (×200). Scale bars, 50 µm. b. miR-494 mimics inhibited G0/G1 to S phase progression in the cell cycle in HTR-8/SVneo cells. c. miR-494 mimics inhibited HTR-8/SVneo proliferation, as shown by CCK-8 assays. d. miR-494 mimics upregulated HTR-8/SVneo P21 and P16 protein levels, as shown by WB. The data are presented as the mean ± SD from three independent experiments, and every experiment was repeated three times, *p < 0.05, **p < 0.01, ***p < 0.001.
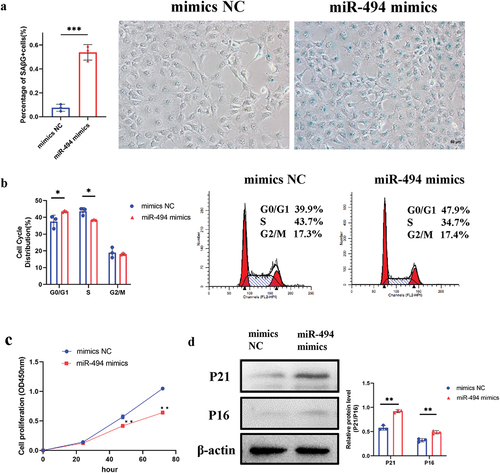
Figure 4. MiR-494 decreased the viability and increased the ROS and inflammatory molecule production of HTR-8/SVneo cells a. miR-494 mimics inhibited HTR-8/SVneo cell migration (×100). Scale bars, 100 µm. b. miR-494 mimics increased HTR-8/SVneo cell ROS levels (×200). Scale bars, 200 µm. c. miR-494 mimics decreased HTR-8/SVneo cell ATP levels. d. miR-494 mimics increased HTR-8/SVneo NLRP3 and IL-1β protein levels, as shown by WB. The data are presented as the mean ± SD from three independent experiments, and every experiment was repeated three times, *p < 0.05, **p < 0.01, ***p < 0.001.
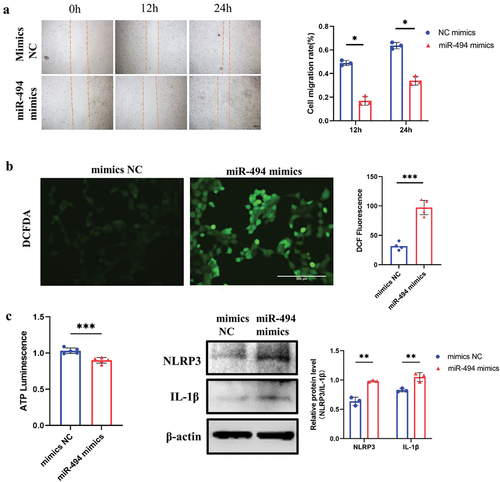
MiR-494 decreases HTR-8/SVneo viability and increases ROS and inflammatory molecule production
To assess the effects of miR-494 overexpression on trophoblast viability, HTR-8/SVneo cells transfected with miR-494 mimics or mimic NC were subjected to migration and ATP assays. The migratory capability was markedly suppressed, as shown in the wound healing assay (, p < 0.05), and ATP synthesis was decreased in miR-494 mimic-transfected cells (, p < 0.001). In addition, the ROS level in miR-494 mimic-transfected cells was 3-fold higher than that in mimic NC-transfected cells (, p < 0.001), and the protein levels of NLRP3 and IL-1β were significantly upregulated (, all p < 0.01).
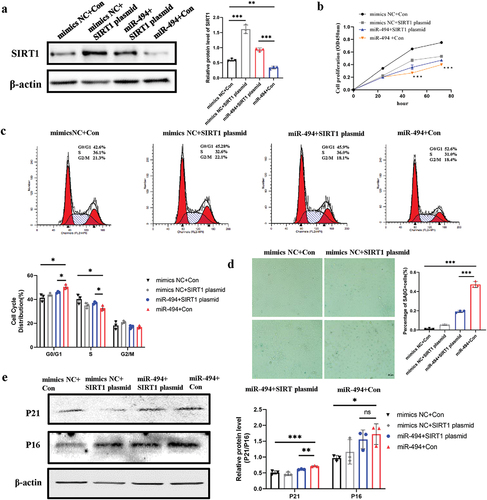
SIRT1 plasmids partially rescued HTR-8/SVneo senescence induced by miR-494 mimics
We conducted a rescue experiment to demonstrate that miR-494 targets SIRT1 and induces trophoblast senescence. HTR-8/SVneo cells were transfected with mimic NC+ the control (Con) plasmid, mimic NC+ the SIRT1 plasmid, miR-494 mimic+ the SIRT1 plasmid, or miR-494 mimics+ the Con plasmid, and SIRT1 protein expression was measured after 48 h of transfection (). In miR-494 mimic-transfected cells, SIRT1 plasmids partially rescued proliferation defects (, p < 0.001) and cell cycle arrest (, p < 0.05). SIRT1 plasmids partially decreased SAβG-positive staining (, p < 0.001) and the protein levels of P21 (, p < 0.01).
Figure 5. SIRT1 plasmids partially rescued HTR-8/SVneo cell senescence induced by miR-494 mimics a. Western blotting was used to measure SIRT1 protein expression in HTR-8/SVneo cells after miR-494 and SIRT1 expression regulation. b. SIRT1 plasmids partially reduced HTR-8/SVneo cell proliferation, as shown by CCK-8 assays. c. SIRT1 plasmids partially rescued the G0/G1-to-S-phase progression of the cell cycle in HTR-8/SVneo cells. d. SIRT1 plasmids partially weakened the activity of SaβG (×200). Scale bars, 50 µm. e. SIRT1 plasmids partially decreased the protein levels of P21 and P16, as shown by WB. The data are presented as the mean ± SD from three independent experiments, and every experiment was repeated three times, *p < 0.05, **p < 0.01, ***p < 0.001.
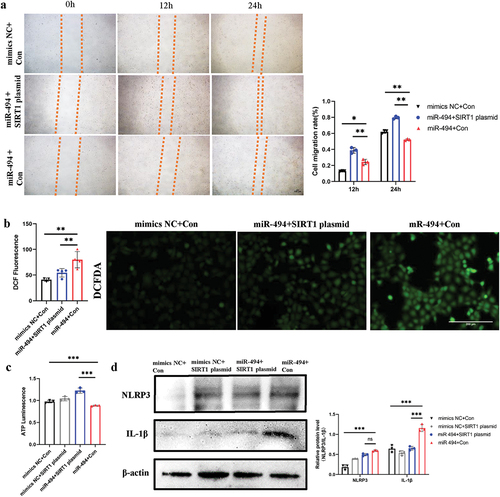
SIRT1 plasmids rescued the viability of HTR-8/SVneo cells and partially decreased the production of ROS and inflammatory molecules
In contrast to miR-494 mimic, SIRT1 plasmids significantly rescued and improved trophoblast migration (, p < 0.01) and ATP synthesis (, p < 0.001). SIRT1 plasmids partially decreased the levels of ROS (, p < 0.01). The levels of the inflammatory molecules NLRP3 and IL-1β (, p < 0.001) showed downward trends, although the NLRP3 level changes were not statistically significant (, p > 0.05).
Figure 6. SIRT1 plasmids rescued HTR-8/SVneo cell viability and partially decreased ROS and inflammatory molecule production a. SIRT1 plasmids reduced HTR-8/SVneo cell migration (×100). Scale bars, 100 µm. b. SIRT1 plasmids partially weakened miR-494 mimic-induced ROS production in HTR-8/SVneo cells (×200). Scale bars, 200 µm. c. SIRT1 plasmids reduced ATP levels in HTR-8/SVneo cells induced by miR-494 mimics. d. SIRT1 plasmids downregulated NLRP3 and IL-1β protein levels in HTR-8/SVneo cells, as shown by WB. The data are presented as the mean ± SD from three independent experiments, and every experiment was repeated three times, *p < 0.05, **p < 0.01, ***p < 0.001.
Discussion
In this study, miR-494 overexpression was shown to be associated with the induction of trophoblast cell senescence and the promotion of inflammation. The possible mechanism might be the direct binding of miR-494 to SIRT1. SIRT1 is a well-known longevity gene. Xiong L (Citation23) confirmed that SIRT1 is involved in the regulation of trophoblast senescence from the first trimester of pregnancy. Yin (Citation24) reported that D-galactose may induce HTR8/SVneo premature aging through the SIRT1/FOXO3a/ROS signaling pathway mediated by oxidative stress and that SIRT1 protects cells from this damage. Wang (Citation16) demonstrated that downregulation of SIRT1 may accelerate placental senescence via targets contributing to the regulation of the cell cycle, ECM production and cytoskeleton reorganization. An imbalance in redox homeostasis could be the mechanism underlying the loss of trophoblast SIRT1 expression in PE (Citation8,Citation25,Citation26). Consistent with previous studies, we found increased numbers of SaβG-positive cells in the placentas of patients with SPE. SIRT1 expression was significantly lower in the SPE group than in the normal group.
Recent experimental data have revealed that placental miRNAs regulate gene networks and affect trophoblast differentiation and angiogenesis. The biological functions of placental miRNAs in PE are becoming evident (Citation19,Citation27); however, few articles (Citation28,Citation29) have reported a role for miRNAs in regulating trophoblast senescence. We combined the results of analyses of several databases and hypothesized that miR-494 targets SIRT1 and potentially participates in placental aging in PE patients. We subsequently found that miR-494 expression was elevated in the placental tissues of PE patients. Dual-luciferase assays revealed that SIRT1 is directly targeted by miR-494. In the cell experiments, miR-494 overexpression decreased SIRT1 mRNA and protein expression, further confirming the relationship between SIRT1 and miR-494.
Several reports have demonstrated the biological roles of miR-494 in human diseases (Citation30,Citation31). In research on pregnancy-related disease, miR-494 has been shown to be the key regulator of endometrial receptivity through the PI3K/AKT/mTOR pathway (Citation32). miR-494 is highly expressed in decidua-derived MSCs (dMSCs) from patients with PE and arrests dMSCs at the G1/S transition by targeting CDK6 and CCND1 (Citation33). Here, we studied the impact of miR-494 on viability and migration in the trophoblast cell line HTR-8/SVneo. Our analyses focused on decreased proliferation, cell cycle arrest, SAβG accumulation and P21/P16 protein activation and suggested that miR-494 overexpression can induce cell senescence. Thus, HTR-8/SVneo senescence induced by miR-494 overexpression likely decreases migration function. However, in the rescue experiment, there was a significant change in P21, while the change in P16 was not statistically significant. We speculated that SIRT1 directly targets the p53/p21 signaling pathway, leading to the aging of cells (Citation34). Decreased ATP levels and increased ROS generation may occur due to decreased SIRT1 antioxidant responses and mitochondrial damage in aging cells (Citation23,Citation35,Citation36). Further experimental evidence is needed.
Interestingly, we also found that overexpression of miR-494 resulted in marked increases in NLRP3 and IL-1β protein levels in HTR-8/SVneo cells, consistent with reports about inflammatory activation in PE (Citation37). We hypothesize that this finding is related to the downregulation of SIRT1 and cell senescence. A rescue experiment suggested that SIRT1 plasmid transfection weakened the HTR-8/SVneo cell senescence phenotype and inhibited NLRP3 and IL-1β production. Although SIRT1 has been shown to regulate the NLRP3 inflammatory body, its potential molecular mechanism remains to be determined. NLRP3 inflammasome activation can be induced by mitochondrial dysfunction, ROS production and lysosomal damage (Citation38,Citation39). SIRT1 can partly inhibit the NLRP3 inflammasome and may be activated by inhibiting oxidative stress and antisenescence.
Conclusion
Overall, we have demonstrated that the miR-494/SIRT1 interaction plays a role in trophoblast senescence and is a possible mechanism underlying premature placental aging in PE patients. This work supplements the existing knowledge regarding the mechanism underlying PE progression and placental aging. Because this work was limited by its small scale, a larger-scale patient cohort should be initiated for further confirmation and advancements. In addition, further studies are needed to determine whether the miR-494/SIRT1 interaction is involved in other PE subtypes.
Informed consent
Informed consent was obtained from all available or living individual participants included in the study.
Data availability statement
The data underlying this article are available in Figshare at https://doi.org/10.6084/m9.figshare.22816079
Additional information
Funding
References
- Abalos E, Cuesta C, Grosso AL, et al. Global and regional estimates of preeclampsia and eclampsia: a systematic review. Eur J Obstet Gynecol Reprod Biol. 2013;170:1–12.
- Susan JF. Why is placentation abnormal in preeclampsia? Am J Obstet Gynecol. 2015;213(4):S115–22.
- Apicella C, Ruano CSM, Méhats C, et al. The role of epigenetics in placental development and the etiology of preeclampsia. Int J Mol Sci. 2019;20(11):2837.
- Holland O, Dekker Nitert M, Gallo LA, et al. Review: placental mitochondrial function and structure in gestational disorders. Placenta. 2017;54:2–9.
- Sultana Z, Maiti K, Aitken J, et al. Oxidative stress, placental ageing-related pathologies and adverse pregnancy outcomes. Am J Reprod Immunol. 2017;77(5):e12653.
- Hernandez-Segura A, Nehme J, Demaria M. Hallmarks of cellular senescence. Trends Cell Biol. 2018;28(6):436–453.
- Cindrova-Davies T, Fogarty NME, Jones CJP, et al. Evidence of oxidative stress induced senescence in mature, post-mature and pathological human placentas. Placenta. 2018;68:15–22.
- Paula JS, Simpson A, Kurlak LO, et al. Increased placental cell senescence and oxidative stress in women with pre-eclampsia and normotensive post-term pregnancies. Int J Mol Sci. 2021;22(14):7295.
- Siddique N, Cox B. Computational analysis identified accelerated senescence as a significant contribution to preeclampsia pathophysiology. Placenta. 2022;121:70–78.
- Cox LS, Redman C. The role of cellular senescence in ageing of the placenta. Placenta. 2017;52:139–145.
- Sultana Z, Maiti K, Dedman L, et al. Is there a role for placental senescence in the genesis of obstetric complications and fetal growth restriction? Am J Obstet Gynecol. 2018;218(2):S762–S773.
- Chen C, Zhou M, Ge Y, et al. SIRT1 and aging related signaling pathways. Mech Ageing Dev. 2020;187:111215.
- Singh CK, Chhabra G, Ann Ndiaye M, et al. The role of sirtuins in antioxidant and redox signaling. Antioxid Redox Signal. 2018;28(8):643–661.
- Liu Z, Wang C, Pei J, et al. SIRT1: a novel protective molecule in pre-eclampsia. Int J Med Sci. 2022;19(2):993–1002.
- Lee KM, Seo HW, Kwon MS, et al. SIRT1 negatively regulates invasive and angiogenic activities of the extravillous trophoblast. Am J Reprod Immunol. 2019;82(4):e13167.
- Wang Y, Zhang Y, Wu Y, et al. SIRT1 regulates trophoblast senescence in premature placental aging in preeclampsia. Placenta. 2022;122:56–65.
- Olivieri F, Rippo MR, Monsurro V, et al. MicroRNAs linking inflamm-aging,cellular senescence and cancer. Ageing Res Rev. 2013;12(4):1056–1068.
- Majidinia M, Mir SM, Mirza-Aghazadeh-Attari M, et al. MicroRNAs, DNA damage response and ageing. Biogerontol. 2020;21(3):275–291.
- Vaiman D. Genes, epigenetics and miRNA regulation in the placenta. Placenta. 2017;52:127–133.
- Varghese F, Bukhari AB, Malhotra R, et al. IHC profiler: an open source plugin for the quantitative evaluation and automated scoring of immunohistochemistry images of human tissue samples. PLoS ONE. 2014;9(5):e96801.
- Jensen EC. Quantitative analysis of histological staining and fluorescence using ImageJ. Anat Rec. 2013;296(3):378–381.
- Lozano-Gerona J, García-Otín ÁL. ImageJ-based semiautomatic method to analyze senescence in cell culture. Anal Biochem. 2018;543:30–32.
- Xiong L, Ye X, Chen Z, et al. Advanced maternal age-associated SIRT1 deficiency compromises trophoblast epithelial-mesenchymal transition through an increase in vimentin acetylation. Aging Cell. 2021;20(10):e13491.
- Yin L, Xu L, Chen B, et al. SRT1720 plays a role in oxidative stress and the senescence of human trophoblast HTR8/SVneo cells induced by D-galactose through the SIRT1/FOXO3a/ROS signalling pathway. Reprod Toxicol. 2022;111:1–10.
- Manna S, McCarthy C, McCarthy FP. Placental ageing in adverse pregnancy outcomes: telomere shortening, cell senescence, and mitochondrial dysfunction. Oxid Med Cell Longev. 2019;2019:3095383.
- Alves-Fernandes DK, Jasiulionis MG. The role of SIRT1 on DNA damage response and epigenetic alterations in cancer. Int J Mol Sci. 2019;20(13):3153.
- Bounds KR, Chiasson VL, Pan LJ, et al. MicroRNAs: new players in the pathobiology of preeclampsia. Front Cardiovasc Med. 2017;4:60.
- Saenen ND, Martens DS, Neven KY, et al. Air pollution-induced placental alterations: an interplay of oxidative stress, epigenetics, and the aging phenotype. Clin Epigenet. 2019;11:124.
- Tsamou M, Martens DS, Cox B, et al. Sex-specific associations between telomere length and candidate miRNA expression in placenta. J Transl Med. 2018;16:254.
- Weng JH, Yu CC, Lee YC, et al. MiR-494-3p induces cellular senescence and enhances radiosensitivity in human oral squamous carcinoma cells. Int J Mol Sci. 2016;17(7):1092.
- Zeng Q, Zeng J. Inhibition of miR-494-3p alleviates oxidative stress-induced cell senescence and inflammation in the primary epithelial cells of COPD patients. Int Immunopharmacol. 2021;92:107044.
- Yuan L, Feng F, Mao Z, et al. Regulation mechanism of miR-494-3p on endometrial receptivity in mice via PI3K/AKT/mTOR pathway. Gen Physiol Biophys. 2021;40:351–363.
- Chen S, Zhao G, Miao H, et al. MicroRNA-494 inhibits the growth and angiogenesis regulating potential of mesenchymal stem cells. FEBS Lett. 2015;589:710–717.
- Chen C, Zhou M, Ge Y, et al. SIRT1 and aging related signaling pathways. Mech Ageing Dev. 2020;187:111215.
- Kauppila TES, Kauppila JHK, Larsson NG. Mammalian mitochondria and aging: an update. Cell Metab. 2017;25:57–71.
- Shpilka T. Haynes CM.The mitochondrial UPR: mechanisms, physiological functions and implications in ageing. Nat Rev Mol Cell Biol. 2018;19:109–120.
- Michalczyk M, Celewicz A. The role of inflammation in the pathogenesis of preeclampsia. Mediators Inflamm. 2020;2020:3864941.
- Harijith A, Ebenezer DL, Natarajan V. Reactive oxygen species at the crossroads of inflammasome and inflammation. Front Physiol. 2014;5:352.
- Kelley N, Jeltema D, Duan Y, et al. The NLRP3 inflammasome: an overview of mechanisms of activation and regulation. Int J Mol Sci. 2019;20(13):3328.