ABSTRACT:
Aspergillus subgenus Nidulantes with nine section forms the second largest subgenus of the fungi that comes under the genus Aspergillus. Species in this group of fungi are important as they are reported to play several important roles in the environment including influencing air quality in confined spaces, food spoilage, production of mycotoxins as well as in human pathogenicity. In the present study, 53 strains of Aspergillus subgenus Nidulantes (section: Nidulantes & Usti) isolated from Korea and preserved at the Korean Agricultural Culture Collection (KACC) were subjected to re-identification by using a combined dataset of partial β-tubulin (BenA), Calmodulin (CaM) gene sequences as well as their morphological data. We confirmed 14 species from 53 isolates in Korea. Of them, eleven species were reported in Korea previously (A. amoenus, A. baeticus, A. calidoustus, A. creber, A. insuetus, A. jensenii, A. nidulans, A. protuberus, A. sydowii, A. tabacinus and A. unguis), and three species (A. griseoaurantiacus, A. puulaauensis and A. sublatus) were previously unreported from Korea. We detailed the characteristic features of these three species, that remain unexplored in Korea.
1. Introduction
Aspergillus is a genus of cosmopolitan fungi and belongs to the family Aspergillaceae. It currently contains approximately 446 species, with several species drawing human interest in fields such as biotechnology, human health and the food industry [Citation1,Citation2]. The continuous emergence of new species within this genus reveals its high biodiversity among other fungi. Owing to their varied characteristics, the strains of this genus can grow on diverse substrates, and occur in nature as endophytes, saprophytes, parasites, food contaminants, and human pathogens [Citation2–5].
The subgenus Nidulantes is found to be the second largest subgenus of Aspergillus next to Circumdati and comprises of nine sections, twenty-three series with around 130 species [Citation6]. Most of the species in this subgenus exhibit several characteristic traits including having biseriate conidial heads, conidiophores to be brown-pigmented harboring globose and echinulate type conidia. Initially, in the subgenus Nidulantes five sections namely Versicolores, Nidulantes, Terrei, Usti and Flavipedes, were established based on morphological structures [Citation7]. Later, members of this subgenus underwent phylogenetic re-analysis using DNA sequences of their internal transcribed spacer (ITS) region, β-tubulin gene (BenA), Calmodulin gene (CaM), RNA polymerase II gene (RPB2), and the large subunit 28S ribosomal DNA sequences (LSU), to determine infrageneric relationship and were updated to include several new species [Citation8,Citation9]. Subsequently, section Aenei was also introduced in the subgenus Nidulantes based on emergent phylogenetic analyses which led to inclusion of several additional species [Citation10]. A polyphasic phylogenetic approach led to defining nine sections in Nidulantes, which resulted in addition of a new section, Cavernicolarum [Citation6]. Houbraken et al. provided an extensive overview of families and genera of the Eurotiales which led to the introduction of an updated subgeneric, section as well as series classification [Citation1]. In the recent times, a broad species concept which includes only four Aspergillus species, namely A. creber, A. versicolor, A. sydowii, and A. subversicolor, has been introduced in the series Versicolores [Citation11]. Moreover, in recent times, many species were updated in the subgenus Nidulantes which include, A. qilianyuensis in the section Nidulantes [Citation5]; A. sigarelli in the section Usti [Citation12]; A. sichuanensis and A. tibetensis in the section Aenei [Citation13]; A. hainanicus, A. guangdongensis and A. guangxiensis were described in the sections Cavernicolarum, Ochraceorosei and Sparsi respectively [Citation5, Citation13]. A new series Hainanici was proposed in the section Cavernicolarum to accommodate A. hainanicus [Citation5].
In the Korean Agricultural Culture Collection (KACC), Aspergillus strains have been deposited since the 1990s and were formerly identified mainly based on their morphological features. Presence of cryptic species has been a significant obstacle as morphology-based identification was frequently found to be ambiguous in the sections of Aspergillus over the last two decades [Citation14]. Therefore, multi-locus sequence analysis is currently in use as a reliable approach, for the identification and phylogeny of Aspergillus [Citation2]. To accurately identify Aspergillus, a polyphasic approach which includes morphological, molecular, as well as ecological analysis, and extrolite profiling has been proposed [Citation15].
In this study, a part of Aspergillus strains preserved at the KACC since 1995–2022 were analyzed using their DNA sequence data as well as their morphological characteristics. The identification of strains was based on the partial β-tubulin (BenA) as well as Calmodulin (CaM) gene sequences of the selected fungal strains. To date, 84 different species of Aspergillus have been identified and described in Korea [Citation16–18] and of them, 14 species are included in subgenus Nidulantes. This study aimed to re-identify fungal strains belonging to Aspergillus subgenus Nidulantes from Korea preserved at KACC and provide a detailed description of the unrecorded species from Korea based on their morphological as well as molecular characteristics in addition to supplementing the existing information on the diversity of Aspergillus species from Korea.
2. Materials and methods
2.1. Strains
A total of 53 Korean strains belonging to the genus Aspergillus subgenus Nidulantes in KACC were studied. The strains studied were isolated from all across Korea. The strains were retrieved from storage in Malt extract broth (BD Difco, Sparks, MD, USA), and later moved to Malt Extract Agar (MEA) [Oxoid, Basingstoke, UK]. Details of the strains examined are listed in the .
Table 1. Aspergillus subgenus Nidulantes strains used in this study.
2.2. DNA extraction, amplification, and sequencing
Cultures were grown on MEA plates and DNeasy® plant mini kit (Qiagen, Hilden, Germany) was used for DNA isolation. The DNA templates were quantified using a NanoDrop Spectrophotometer (Thermo Scientific, Waltham, MA, USA). Partial BenA and CaM genes were amplified following the protocols reported earlier by Glass et al. [Citation19] and Hong et al. [Citation20]. The PCR products were bidirectionally sequenced at Macrogen Inc., South Korea, with the same PCR primers and the raw sequences were assembled using DNA STAR Lasergene SeqMan Pro v.10.0.1. (DNASTAR, Inc. Madison, WI).
2.3. Phylogenetic analyses
The obtained gene sequences were combined with reference (preferably ex-type) sequences that were retrieved from earlier studies [Citation1]. Multiple sequence alignment of respective locus was separately performed using CLUSTAL W program in MEGA 11. The alignments were improved following visual inspection and concatenated later in the same software (MEGA 11). Maximum likelihood (ML) phylogenetic trees were generated based on separate and combined datasets of the gene sequences (BenA and CaM). The trees with 1,000 ultrafast bootstrap replications and aBayes support were generated using IQ-TREE and the substitution model options were auto-evaluated as per the provided alignment files [Citation21]. The sequence of A. cibarius KACC 46346 was used as an outgroup. Details of the reference sequences used have been provided in Supplementary Table 1. Sequences obtained during this study were deposited to RDA-GeneBank (http://genebank.rda.go.kr).
2.4. Phenotypic analysis
The fungal strains were inoculated on four different media namely Malt Extract agar (MEA), Czapek Yeast extract agar (CYA), Dichloran 18% Glycerol agar (DG18), and Yeast extract sucrose agar (YES) [Citation22] and incubated at 25 °C for 7 days. At the end of the incubation period, colony diameters and characteristics were recorded. Fungal slides were then prepared with lactic acid and observed under a Zeiss AXIO Imager A1 microscope with differential interference contrast (DIC) illumination, and a digital AxioCam ICc3 camera (Carl Zeiss, Göttingen, Germany). Characteristics including the size, shape, conidial pigmentation and conidiophores were recorded.
3. Results
3.1. Phylogenetic analyses
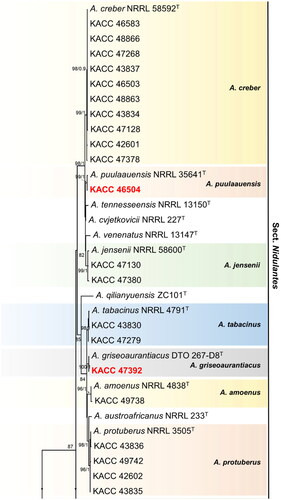
A combined BenA and CaM sequence dataset was used to understand the phylogenetic relationships of KACC strains with other publicly available (ex-) type species belonging to the subgenus Nidulantes (). The concatenated alignment of 74 sequences including our strains as well as (ex-) type species spanning 20 taxa contained 1024 characters (including alignment gaps). Of these, 579 characters were from CaM, and 445 characters from BenA (Supplementary Table 2). The concatenated phylogenetic tree indicated that the 53 strains taken in this study were spread across 14 different Aspergillus species, of which eleven species were earlier reported in Korea, whereas three species (red colored bold text) were hitherto not described from Korea (). Phylogenetic tree was constructed based on single gene were provided in the supplementary figure 1 and 2. During BLAST analysis, we observed the BenA sequence of strain KACC 46504 to express 100% similarity with A. puulaauensis NRRL35641T followed by 99.2% similarity with A. cvjetkovicii NRRL227T (98.94%) and A. jensenii NRRL58600T (98.14%). This was also consistent with the BLAST results of the strain using CaM sequence where the top nearest hits were A. puulaauensis NRRL35641T (99.6%), A. cvjetkovicii NRRL227T (99.4%) and A. tennesseensis NRRL13150T (99.19%). Interestingly, in case of strain KACC 46824, the top BLAST hits for similarity with its BenA sequence were found to be A. sublatus CBS140630T (100%) and A. quadrilineatus NRRL201T (100%). However, BLAST results from CaM sequence analysis of the strain revealed a higher similarity with A. sublatus CBS140630T (99.34%) compared to A. quadrilineatus NRRL201T (98.68%). It also indicated one-hundred percent sequence similarity with another strain of A. sublatus DTO421-D4. To further confirm its identity, we studied the strain KACC 46824 for its macro and micro-morphology. The macromorphology of the strain was consistent with the type strain of A. sublatus/A. latus and in its micromorphology, comparing the vesicle shape indicated its likeness to A. sublatus/A. latus compared to A. quadrilineatus type strain. Finally, BLAST analysis in case of strain KACC 47392 indicated its top hits of BenA sequence to be A. griseoaurantiacus DTO:267-D8T (100%) followed by A. tabacinus NRRL4791T (99.47%) and A. amoenus NRRL4838T (98.93%). This was substantiated by the BLAST of its CaM sequence which showed the highest similarity of the sequence to be with A. griseoaurantiacus DTO:267-D8T (100%) followed by A. tabacinus NRRL4791T (98.99%) and A. amoenus NRRL4838T (98.91%).
Figure 1. Phylogenetic position of Aspergillus subgenus Nidulantes strains from the KACC based on a combined data set containing partial BenA and CaM sequences. Bootstrap values ≥70 (left) and aBayes values ≥0.9 (right) are presented at the nodes. The scale bar indicates the number of substitutions per nucleotide. The unrecorded species are represented in bold & red in color. Ex-type strains are denoted byT. The species A. cibarius was used as the outgroup.
In the section Nidulantes, 50 strains grouped into eleven clusters, with A. amoenus, A. creber, A. griseoaurantiacus, A. jensenii, A. nidulans, A. protuberus, A. puulaauensis, A. sublatus, A. sydowii, A. tabacinus and A. unguis as their nearest neighbors. Until now, 76 species of section Nidulantes have been reported worldwide [Citation1, Citation5]. Among them, A. amoenus, A. creber, A. jensenii, A. nidulans, A. protuberus, A. subversicolor, A. sydowii, A. tabacinus, A. tennesseensis, A. unguis and A. versicolor are recorded in Korea [Citation16, Citation23]. Species A. puulaauensis, A. griseoaurantiacus and A. sublatus were not previously recorded in Korea, and are incorporated now in the present report.
Among 26 known species from the section Usti [Citation1, Citation12]; five of them, viz., A. baeticus, A. calidoustus, A. germanicus, A. insuetus and A. pseudodeflectus have been reported from Korea [Citation16]. Three strains from the section Usti in Korea grouped with type strains of A. calidoustus, A. insuetus and A. baeticus as their closest neighbors and identified respectively.
3.2. Taxonomy
Aspergillus puulaauensis Jurjević, S.W. Peterson & B.W. Horn, IMA Fungus 3 (1): 71 (2012) [MB#800602] [Citation24]
Colony characteristics: The fungal colonies attain 20–21 mm diameter in a span of 7 days at 25 °C on CYA, sulcate with artemisia green conidial heads, no soluble pigment, reverse brown. The colonies were sulcate, centrally raised light sporulation with funicular hyphal clumps, surrounded by yellowish sporulation with white mycelium at periphery, yellowish brown color in reverse and reaches 17-18 mm diameter in 7 days on MEA. The colonies were clear white fine sporulation, white in reverse and extended 15-16 mm in diameter at 25 °C after 7 days on DG18. Irregular colony appearance with artemisia green conidial heads seen on further incubation. Colonies attain 32–33 mm diameter on YES at 25 °C after 7 days; light grayish green sporulation at center followed by yellowish sporulation with white mycelium at margins, reverse pale yellow.
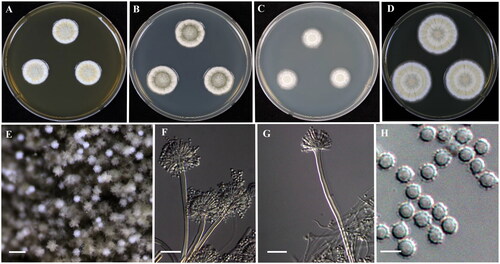
Micromorphology: Presence of biseriate conidial heads, smooth-walled, stipes hyaline, 150–320 × 4–5 μm. Pyriform to spatulate vesicles, 5–11 µm. Metulae covering half to entire surface of the vesicle, 4–6 × 3–4 μm. Flask-shaped phialides, 5–7 × 2–3.5 μm, fragmentary heads resembling penicillate fructifications were occasionally present. Conidia spherical to ellipsoidal, often covered by a thick layer (about 0.3 μm), rough, 2–3 μm ().
Figure 2. Morphology of Aspergillus puulaauensis (KACC 46504). (A–D) Colonies grown on MEA, CYA, DG18 and YES media after 7 days at 25 °C from left to right. (E) Conidial head on MEA, (F,G) Conidiophores with conidial head & (H) Conidia. Scale bars: E = 125 µm, F, G = 25 µm, H = 5 µm.
Examined strain: KACC 46504
Remarks: KACC 46504 was alike A. puulaauensis described by Jurjević et al. [Citation24]. However, conidia were slightly smaller (2–3 μm) than that of A. puulaauensis NRRL 35641 (2.5–5.5 μm) [Citation24]. Moreover, colonies on YES media showed soluble pink pigment after 14 days at 25 °C.
Aspergillus griseoaurantiacus Visagie, Hirooka & Samson, Studies in Mycology 78: 112 (2014) [MB#809197] [Citation22]
Colony characteristics: In medium CYA, the colonies were floccose, mycelial areas white to light brown with bluish green sporulation, reverse brown and reached 16–17 mm diameter after 7 days at 25 °C. Colonies on MEA were found to be floccose, dark bluish green with dull brown sporulation at center, encircled by white mycelium at margins, exudate minute brown droplets on further incubation of the colony, reverse yellow and attains 14–15 mm diameter at 25 °C after 7 days. Colonies were white in appearance, slow growth, reverse yellow and extended 7-8 mm in diameter at the end of 7 days at 25 °C on DG18. On further incubation, colonies exhibited artemisia green conidial heads and irregular colony margin. Colonies on YES attain 26–27 mm diameter after 7 days at 25 °C; Colony surface floccose, moderately radially sulcate toward the center, mycelium white, light greenish ash sporulation, reverse yellow.
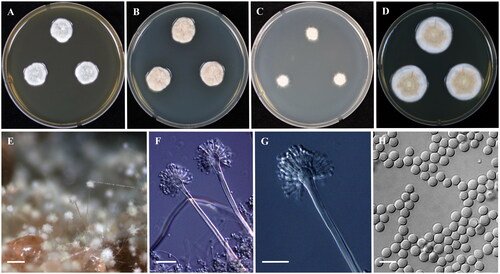
Micromorphology: The fungal conidial heads were radiating biseriate, smooth-walled, stipes hyaline, 130–350 × 4–6 μm. Vesicles spatulate, 10–16 µm. Metulae covering 85% of the head, 4–7 × 3–4 μm. Ampulliform phialides, 5–7 × 2.5–3.5 μm. Conidia were found to be globose to ellipsoidal and smooth, 2–3 μm ().
Figure 3. Morphology of Aspergillus griseoaurantiacus (KACC 47392). (A-D) Colonies grown on MEA, CYA, DG18 and YES media after 7 days at 25 °C from left to right. (E) Conidial head on MEA, (F,G) Conidiophores with conidial head & (H) Conidia. Scale bars: E = 125 µm, F, G = 25 µm, H = 5 µm.
Examined strain: KACC 47392
Remarks: KACC 47392 was morphologically close to A. griseoaurantiacus earlier described by Visagie et al. [Citation22]. However, KACC 47392 showed globose to ellipsoidal, smooth conidia but A. griseoaurantiacus CBS 138191 has ellipsoidal, finely roughened conidia [Citation22]. In addition, colonies on YES media, indicated minute brown droplets like exudate after 14 days at 25 °C.
Aspergillus sublatus Y. Horie, Transactions of the Mycological Society of Japan 20: 481 (1979) [MB#118407] [Citation25]
Colony characteristics: The colonies were moderately deep with white mycelium, no soluble pigment, light brown sporulation, reverse brown and eventually reaching a diameter of 36-37 mm at the end of 7 days at 25 °C on CYA medium. The surface of the colonies was found to be floccose with white mycelial areas and light brown to green sporulation, no soluble pigment, brownish-yellow in reverse, and further reached 30–32 mm in diameter on MEA at 25 °C after 7 days. On DG18, white colony mycelium and sporulation green in color and white in reverse with 10–12 mm in diameter at 25 °C after 7 days. On YES medium, the colonies were floccose, white mycelial areas, wrinkled with 58–60 mm in diameter after 7 days at 25 °C; conidia green, reverse wrinkled, yellowish orange that faded into light yellow toward the margins.
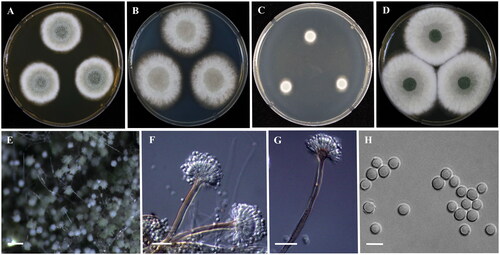
Micromorphology: Microscopic observation found the conidial heads to be biseriate and at instances reduced Penicillium-like structures were present, Brown hyaline stipes, smooth-walled, 90-250 × 4–6 µm. Vesicles were subglobose to subclavate, brown, measuring 8–11 µm and covered the upper half of the head; Metulae 3–7 × 2–4 μm. Phialides were flask-shaped, 5–9 × 2–3.5 μm. Conidia were observed to be globose to subglobose, smooth, 3–4 μm ().
Figure 4. Morphology of Aspergillus sublatus (KACC 46824). (A–D) Colonies grown on MEA, CYA, DG18 and YES media after 7 days at 25 °C from left to right. (E) Conidial head on MEA, (F,G) Conidiophores with conidial head & (H) Conidia. Scale bars: E = 125 µm, F, G = 25 µm, H = 5 µm
Examined strain: KACC 46824
Remarks: A. sublatus was synonymized with A. latus [Citation6]. KACC 46824 showed excellent growth at 37 °C on CYA with dense sporulation. Green sporulation was enhanced on colonies after incubation of 14 days at 25 °C.
4. Discussion
In the current study, we aimed at re-identifying the strains previously submitted to KACC as Aspergillus or its earlier related sexual state genera (e.g. Emericella), based on a combination of molecular and morphological data. In earlier days, morphological characteristics alone were used for identification of these fungal strains and then ITS was recognized as a universal DNA marker for a more accurate identification of fungi. However, earlier studies have shown that the ITS sequence was insufficient for the accurate identification of Aspergillus species and recommended BenA and CaM gene as the suitable markers [Citation1, Citation22]. In the current study, BenA and CaM gene based phylogeny analysis led us to recognize three previously undescribed species of Korea. Until now, only few reports have been made on unrecorded Aspergillus species from Korea despite the ubiquitous distribution. However, there has also been an increase in the reports of several new Aspergillus species throughout the world [Citation13, Citation17].
The previously unrecorded species of Korea, KACC 46504, KACC 47392 and KACC 46824 strains belonged to the section Nidulantes (). Aspergillus section Nidulantes species are ubiquitous in the environment and are believed to perform significant roles in everyday life of humans [Citation6]. Recently, Aspergillus section Nidulantes underwent a taxonomic revision by means of a polyphasic approach, and a series classification was introduced. The revised section harbors 67 species and seven series [Citation1]. In the present study, the three unrecorded species were found to be associated with the Versicolores (two species) and Nidulantes (one species) series. The fungi belonging to the series Versicolores are often mentioned to be as ubiquitous as they are frequently isolated from a wide range of environmental niches including soil, indoors, foods, animal feed, plant sources, caves, and even from clinical material [Citation24, Citation26]. In our case, A. sublatus, A. puulaauensis and A. griseoaurantiacus mainly originated from food and air. The initial report describing A. griseoaurantiacus indicated the species to be present in house dust from various indoor environments of Mexico and Micronesia [Citation22]. Moreover, A. griseoaurantiacus has also been reported to produce the enzyme chitinase and the mycotoxin, sterigmatocystin, making it relevant to biotechnology as well as human health [Citation11, Citation27]. A. puulaauensis however, has been isolated from a comparatively wider spectrum of ecological niches including dead hardwood branch, clinical areas, mold damaged homes, grapes and once from indoor air [Citation24, Citation28]. Previous reports of A. puulaauensis have recorded production of norsolorinic acid, Versicolorin A, 5-methoxyterigmatocystin & sterigmatocystin and also reported cellular cytotoxicity against A549 and HaCaT cell lines [Citation6, Citation28, Citation29]. Few studies showed that A. puulaauensis extrolites to have a potential to be used as a ingredient in cosmetics or used to lower oxidative stress [Citation30]. A. sublatus was treated as a synonym of A.latus [Citation6] but failed to have any priority over A. sublatus. Later, fungal taxonomists considered that the correct name for this species is A. sublatus as prosposed by Y. Horie [Citation25]. Several extrolites such as asperthecin, asperugins, emericellin, shamixanthones, sterigmatocystin, Nidulalin A & B, an emindol, a violaceol and versicolorins production have been reported in A. sublatus [Citation6]. Moreover, A. sublatus is a well-known etiological agent of aspergillosis [Citation13].
The second largest section in subgenus Nidulantes is Usti which encompasses 26 species and 4 series [Citation1, Citation12]. Most fungi from the section Usti, are found to produce close to brown-pigmented conidiophores with coarsely roughened conidia [Citation12, Citation31]. These group of aspergilli have an history of being isolated from cave sediments, indoor environments, soil as well as clinical samples [Citation12, Citation32]. Many species of the section Usti especially A. calidoustus and A. insuetus have been recognized as causal agent of Invasive Aspergillosis [Citation32].
To date, fourteen species have been reported from Korea in the subgenus Nidulantes. The KACC maintains 13 species from several provinces, and most of them originated from soil, air, water, food and clinical samples. In this study, we have described three more unrecorded species in the subgenus Nidulantes which now extends the count to 17 species in Korea. The results of this study will improve knowledge on the distribution of Aspergillus species in Korea and promote development of applications of the various species in the subgenus Nidulantes.
Supplemental Material
Download MS Word (781.5 KB)Disclosure statement
The authors pronounce that they have no potential conflict of interest.
Additional information
Funding
References
- Houbraken J, Kocsubé S, Visagie CM, et al. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): an overview of families, genera, subgenera, sections, series and species. Stud Mycol. 2020;95:5–169. doi: 10.1016/j.simyco.2020.05.002.
- Samson RA, Visagie CM, Houbraken J, et al. Phylogeny, identification and nomenclature of the genus Aspergillus. Stud Mycol. 2014;78(1):141–173. doi: 10.1016/j.simyco.2014.07.004.
- Houbraken J, de Vries RP, Samson RA. Modern taxonomy of biotechnologically important Aspergillus and Penicillium species. Adv Appl Microbiol. 2014;86:199–249. doi: 10.1016/B978-0-12-800262-9.00004-4.
- Frisvad JC, Larsen TO. Chemodiversity in the genus Aspergillus. Appl Microbiol Biotechnol. 2015;99(19):7859–7877. doi: 10.1007/s00253-015-6839-z.
- Wang X-C, Zhuang W-Y. New species of Aspergillus (Aspergillaceae) from tropical islands of China. JoF. 2022;8(3):225. doi: 10.3390/jof8030225.
- Chen AJ, Frisvad JC, Sun BD, et al. Aspergillus section Nidulantes (formerly Emericella): polyphasic taxonomy, chemistry and biology. Stud Mycol. 2016;84(1):1–118. doi: 10.1016/j.simyco.2016.10.001.
- Gams W, Christensen M, Onions AH, et al. Infrageneric taxa of Aspergillus. In Advances in Penicillium and Aspergillus systematics. Edited by Samson RA, Pitt JI. Boston, MA: Springer US; 1986: 55–62.
- Peterson SW. Phylogenetic analysis of Aspergillus species using DNA sequences from four loci. Mycologia. 2008;100(2):205–226. doi: 10.3852/mycologia.100.2.205.
- Peterson SW, Varga J, Frisvad JC, et al. Phylogeny and subgeneric taxonomy of Aspergillus. In: Varga J, Samson RA, editors. Aspergillus in the genomic era. Wageningen: Wageningen Academic Publishers; 2008. p. 33–56.
- Varga J, Frisvad JC, Samson RA. Aspergillus sect. Aeni sect. nov., a new section of the genus for A. karnatakaensis sp. nov. and some allied fungi. IMA Fungus. 2010;1(2):197–205. doi: 10.5598/imafungus.2010.01.02.13.
- Sklenář F, Glässnerová K, Jurjević Ž, et al. Taxonomy of Aspergillus series Versicolores: species reduction and lessons learned about intraspecific variability. Stud Mycol. 2022;102(1):53–93. doi: 10.3114/sim.2022.102.02.
- Sun BD, Houbraken J, Frisvad JC, et al. New species in Aspergillus section Usti and an overview of Aspergillus section Cavernicolarum. Int J Syst Evol Microbiol. 2020;70(10):5401–5416. doi: 10.1099/ijsem.0.004425.
- Sun B, Luo C, Bills GF, et al. Four new species of Aspergillus subgenus Nidulantes from China. JoF. 2022;8(11):1205. doi: 10.3390/jof8111205.
- Balajee SA, Nickle D, Varga J, et al. Molecular studies reveal frequent misidentification of Aspergillus fumigatus by morphotyping. Eukaryot Cell. 2006;5(10):1705–1712. doi: 10.1128/EC.00162-06.
- Stengel A, Stanke KM, Quattrone AC, et al. Improving taxonomic delimitation of fungal species in the age of genomics and phenomics. Front Microbiol. 2022;13:847067. doi: 10.3389/fmicb.2022.847067.
- National List of Species of Korea. National Institute of Biological Resources; 2022.
- Pangging M, Nguyen TTT, Lee HB. Seven undescribed Aspergillus species from different niches in Korea. Mycobiology. 2022;50(4):189–202. doi: 10.1080/12298093.2022.2116158.
- Anbazhagan Mageswari YC, Thao LD, Lee D, et al. Re-identification of Aspergillus subgenus Circumdati strains in Korea led to the discovery of three unrecorded species. Mycobiology. 2023;51(5):288–299. doi: 10.1080/12298093.2023.2257997.
- Glass NL, Donaldson GC. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol. 1995;61(4):1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995.
- Hong SB, Go SJ, Shin HD, et al. Polyphasic taxonomy of Aspergillus fumigatus and related species. Mycologia. 2005;97(6):1316–1329. doi: 10.3852/mycologia.97.6.1316.
- Nguyen LT, Schmidt HA, von Haeseler A, et al. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32(1):268–274. doi: 10.1093/molbev/msu300.
- Visagie CM, Hirooka Y, Tanney JB, et al. Aspergillus, Penicillium and Talaromyces isolated from house dust samples collected around the world. Stud Mycol. 2014;78(1):63–139. doi: 10.1016/j.simyco.2014.07.002.
- Lee YN, Kim NJ, Kang DS. Aspergillus itaconicus and Aspergillus unguis with new addition to the korean flora. Korean J Microbiol. 1977;15(1):1–8.
- Jurjevic Z, Peterson SW, Horn BW. Aspergillus section Versicolores: nine new species and multilocus DNA sequence based phylogeny. IMA Fungus. 2012;3(1):59–79. doi: 10.5598/imafungus.2012.03.01.07.
- Horie Y. New or interesting Emericella from herbal drugs. Trans Mycol Soc Japan. 1979;20:481–491.
- Nováková A, Hubka V, Valinová Š, et al. Cultivable microscopic fungi from an underground chemosynthesis-based ecosystem: a preliminary study. Folia Microbiol (Praha). 2018;63(1):43–55. doi: 10.1007/s12223-017-0527-6.
- Jakšić Despot D, Kocsubé S, Bencsik O, et al. New sterigmatocystin-producing species of Aspergillus section Versicolores from indoor air in Croatia. Mycol Progress. 2017;16(1):63–72. doi: 10.1007/s11557-016-1250-4.
- Géry A, Basset B, Gounel N, et al. Aflatoxin biosynthetic pathway extrolites in airborne Aspergilli series Versicolores. WMJ. 2023;16(2):127–135. doi: 10.3920/WMJ2022.2809.
- Géry A, Lepetit C, Heutte N, et al. Cellular cytotoxicity and oxidative potential of recurrent molds of the genus Aspergillus series Versicolores. Microorganisms. 2022;10(2):228. doi: 10.3390/microorganisms10020228.
- Letsiou S, Bakea A, Goff GL, et al. In vitro protective effects of marine-derived Aspergillus puulaauensis TM124-S4 extract on H2O2-stressed primary human fibroblasts. Toxicol in Vitro. 2020;66:104869. doi: 10.1016/j.tiv.2020.104869.
- Samson RA, Varga J, Meijer M, et al. New taxa in Aspergillus section Usti. Stud Mycol. 2011;69(1):81–97. doi: 10.3114/sim.2011.69.06.
- Glampedakis E, Cassaing S, Fekkar A, et al. Invasive Aspergillosis due to Aspergillus section Usti: a multicenter retrospective study. Clin Infect Dis. 2021;72(8):1379–1385. doi: 10.1093/cid/ciaa230.