Abstract
The fungal strain designated as KNUF-21-020, belonging to the genus Triangularia, was isolated from a soil sample collected in the Chungnam province, Korea. Phylogenetic analyses based on the concatenated nucleotide sequences of internal transcribed spacer regions and partial sequences of large subunit rRNA, beta-tubulin, and RNA polymerase II subunit genes revealed that the strain was grouped in a clade with Triangularia species. However, it occupied a distinct phylogenetic position. We also observed morphological differences between strain KNUF-21-020 and closely related species. Here, we provided detailed descriptions, illustrations, and discussions regarding the morphological and phylogenetic analyses of the closely related species to support the novelty of this isolated species. The phylogenetic analyses and morphological observations indicate that the strain KNUF-21-020 represents a novel species in the genus Triangularia (family: Podosporaceae). We have designated this species as Triangularia manubriata sp. nov.
1. Introduction
Soil-inhabiting fungi play a vital ecological role as decomposers, producing a variety of enzymes that break down organic matter and regulate nutrient balance. Additionally, these important ecosystem regulators strongly influence plant diversity and productivity [Citation1]. The family Podosporaceae was introduced by Wang et al. [Citation2]. It includes three redefined genera, namely, Cladorrhinum, Podospora, and Triangularia, all of which were previously classified in the family Lasiosphaeriaceae. Generally, species in the Podosporaceae family are plant endophytes or saprobes found on rotting wood [Citation2]. These three genera produce single-cell pigmented ascospores or double-cell ascospores comprising a pigmented upper cell and a smaller, usually hyaline, lower cell. The genus Triangularia is typically found in terrestrial habitats, often growing as saprophytes on the ground among leaf litter or in association with plant roots [Citation3]. Morphologically, it is characterized by the specific shape of its double-celled sexual spores, as described above [Citation2,Citation4]. There are currently 23 species in the genus Triangularia, and the type species is Triangularia bambusae [Citation3]. Since Triangularia species have been reclassified from other genera, including Sordaria, Apiosordaria, Cercospora, and Zopfiella, this genus is phylogenetically confusing [Citation4]. Furthermore, several species produce bioactive compounds, such as anserinone A and B, dethiosecoemestrin, and emestrin [Citation5].
This study aimed to expand our knowledge of fungal species diversity in Korea and the use of fungal resources in various industries. Here, we present the morphological and molecular characteristics of strain KNUF-21-020, belonging to the family Podosporaceae.
2. Materials and methods
2.1. Collection of soil samples and fungal strain isolation
Fungal strains were isolated from soil samples collected from Unsan-myeon, Seosan-si, Chungnam province, South Korea (36°46′41.0″N, 126°36′15.8″E). Isolation was performed using the plate dilution method as previously described [Citation6]. Strains KNUF-21-020 and KNUF-21-021 were selected from numerous fungal strains for further morphological and molecular phylogenetic analysis.
2.2. Cultural and morphological characteristics
Strains KNUF-21-020 and KNUF-21-021 were cultured on potato dextrose agar (PDA; Difco, Detroit, MI), malt extract agar (MEA; Difco, Detroit, MI), oatmeal agar (OA; Difco, Detroit, MI), and potato carrot agar (PCA; potato, 20 g; carrot, 20 g; agar, 20 g; distilled H2O, 1000 mL) to study its morphology and growth [Citation3]. The cultural characteristics, including color, shape, and size were recorded after seven days. Morphological characteristics were observed under a BX-50 microscope (Olympus, Tokyo, Japan) and the observed characteristics of both strains were identical.
2.3. Genomic DNA extraction, polymerase chain reaction (PCR) amplification, and sequencing
Due to the identical characteristics of both strains, out of both, only KNUF-21-020 was selected for phylogenetic analysis. We extracted genomic DNA from the mycelia of strain KNUF-21-020 cultured on PDA using the HiGene Genomic DNA Prep Kit (BIOFACT, Daejeon, South Korea) according to the manufacturer’s instructions. Molecular identification was conducted by analyzing sequences of the internal transcribed spacer (ITS) regions and large subunit (LSU) rRNA, beta-tubulin (TUB2), and RNA polymerase II subunit (RPB2) genes, which were amplified separately using the primer pairs ITS1F/ITS4, LROR/LR5, T1/Bt2b, and RPB2-5F/RPB2AM-7R, respectively [Citation7–10]. The amplified PCR products were purified with the EXOSAP-IT PCR Product Cleanup Reagent (Thermo Fisher Scientific, Waltham, MA) and sequenced by SolGent (Daejeon, South Korea). The obtained KNUF-21-020 sequence was deposited in the National Center for Biotechnology Information (NCBI) GenBank database ().
Table 1. GenBank accession numbers of the sequences used for phylogenetic analyses in this study.
2.4. Phylogenetic analysis
Sequences obtained from the NCBI were used to construct phylogenetic relationships (). The ITS region sequences and partial sequences of LSU, TUB2, and RPB2 gene were analyzed for identification supported by the construction of phylogenetic trees based on the neighbor-joining method as described by the Kimura model, using MEGA version X with bootstrap values based on 1000 replications [Citation11–14].
3. Results
3.1. Taxonomy
The morphology of strains KNUF-21-020 and KNUF-21-021 was distinct from the other allied species of Triangularia. Therefore, it was described as a new species. However, since the cultural and morphological characteristics of both KNUF-21-020 and KNUF-21-021 were identical, the strain KNUF-21-020 was selected for analysis.
Triangularia manubriata S. K. Lim, S.Y. Lee, and H.Y. Jung sp. nov. ().
Figure 1. Cultural and morphological characteristics of Triangularia manubriata KNUF-21-020. Cultures were grown at 25 °C for seven days. (A–D) Front and reverse view of the colony on PDA, OA, MEA, and PCA, respectively; (E, F) ascomata produced on PDA; (G) asci; (H) premature (hyaline) and matured (melanized) ascospore. Scale bars: E, F = 50 µm, G, H = 20 µm.
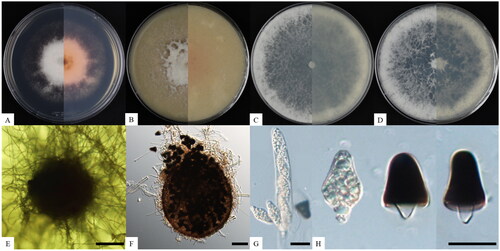
MycoBank: 851263
Etymology: The specific epithet is derived from the Latin word manubrium (“handle”), which refers to the shape of the lower cell of the ascospore.
Typus: The strain was isolated from soil containing plant debris collected from Seosan-si, Chungnam province, South Korea (36°46′41.0″N, 126°36′15.8″E) in 2021. The stock culture was deposited in the National Institute of Biological Resources (NIBR) as a metabolically inactive culture (NIBRFGC000509195).
Habitat: The novel species proposed in this study, Triangularia manubriata, was isolated from soil collected in Korea.
Description: Colonies on PDA reached 63–65 mm in diameter after seven days of culture at 25 °C. They appeared white, dense, and round with abundant aerial mycelia in the center and a flat edge with pink pigmentation on the media. The reverse side showed shades from pink to dark brown (). On OA, the colonies reached up to 80 mm in diameter, and the surface was white, floccose, and circular with entire margins (). On MEA, the colonies reached up to 90 mm after six days, and the surface was white, circular, and floccose, with entire margins on the edge of the mycelium and flat in the center (). Colonies on PCA were similar in size and appearance to colonies on MEA, but were slightly buff in the center (). The ascomata were superficial, dark brown to black under reflected light due to the mass of released ascospores, solitary to aggregated, nonostiolate, and 54–115 μm in diameter (n = 10) (). The asci were cylindrical, containing eight hyaline ascospores that sometimes persisted until the ascospores were mature (). The ascospores were double-celled, 12.0–20.2 × 15.3–25.9 µm long (average: 16.5–20.1, n = 50), and composed of an upper triangular cell with an obtuse end and a handle-shaped hyaline lower cell. The upper triangular cell was hyaline when it matured, at which point the color changed from dark brown to black (). Asexual morphs were absent.
Notes: A comparison of strain KNUF-21-020 with phylogenetically related strains T. bambusae CBS 325.33T and T. setosa CBS 311.58 revealed morphological differences. Strain KNUF-21-020 produced double-celled ascospores composed of a triangular upper cell and a handle-shaped lower cell. T. bambusae and T. setosa also produced double-celled ascospores, but they consisted of an olivaceous brown to dark upper cell and a hyaline lower cell [Citation2] and were oblong shaped. Additionally, KNUF-21-020 ascospores (12.0–20.2 × 15.3–25.9 µm) were smaller than those of T. bambusae (17.0–20.5 × 10.5–12.0 µm) and T. setosa (15.5–16.0 × 18.5–20.0 µm). Thus, the strains were morphologically unique in this genus by the shape and size of their ascospores, specifically the lower cells (). Additionally, its habitat is similar with other Triangularia species, that most species from this genus are isolated from soil or rotting wood that support saprotrophic habitat.
Table 2. Morphological characteristics of Triangularia manubriata (KNUF-21-020T) and comparison with the closest species of Triangularia.
3.2. Phylogenetic analysis
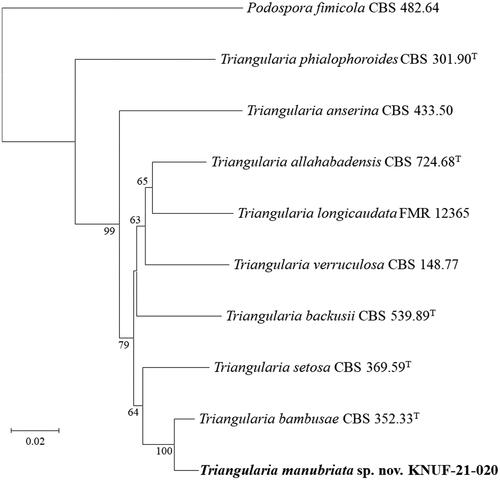
Amplification of the ITS, LSU, RPB2, and TUB2 genes of strain KNUF-21-020 yielded fragments of 391, 747, 1036, and 699 bp, respectively. The ITS regions of strain KNUF-21-020 showed 99.2% similarity with T. bambusae CBS 352.33T and 97.9–98.5% similarity with T. setosa CBS 311.58, T. tetraspora IFO32904, T. unicaudata CBS 313.58, and T. longicaudata FMR 12365. Based on the partial LSU gene sequence, strain KNUF-21-020 showed 100% similarity with T. bambusae CBS 352.33T, and 99.2–99.5% similarity with T. setosa CBS 311.58, T. unicaudata CBS 313.58, and T. longicaudata FMR 12367. Using the partial RPB2 gene sequence, the strain showed a maximum similarity of 96.5% with T. bambusae CBS 352.33T. Similarly, the partial TUB2 gene sequence showed a maximum similarity of 94.5% with T. bambusae CBS 352.33T, and other Triangularia strains showed 89.3–89.7% similarity with T. backusii CBS 539.89, T. unicaudata CBS 313.58, T. longicaudata FMR 12365, and T. allahabadensis CBS 724.68T. Using the four aforementioned molecular markers, all results clearly indicated that comparative analysis based on the sequence of any one gene was insufficient for precise identification of the novel fungal strain at the species level. Therefore, multilocus sequence analysis was performed using concatenated ITS regions and partial LSU, RPB2, and TUB2 gene sequences of strain KNUF-21-020 (). In combination, these four molecular markers were highly effective in species resolution of the genus Triangularia. The NJ phylogenetic tree () based on the concatenated sequences demonstrated that strain KNUF-21-020 occupied a distinct position from other Triangularia species, and its phylogenetically closest neighbor in the genus Triangularia was T. bambusae. Accordingly, the novel strain was considered to represent a single, novel, and phylogenetically distinct Triangularia species.
Figure 2. Neighbor-joining phylogenetic tree based on a combined dataset of the internal transcribed spacer (ITS) regions and partial large-subunit (LSU), beta-tubulin (TUB2), and RNA polymerase II subunit 2 (RPB2) genes sequences showing the phylogenetic position of the strain KNUF-21-020 among Triangularia species. Bootstrap values greater than 60% (percentage of 1000 replications) are shown at branching points. The strain isolated in this study is in bold. The tree was rooted using Podospora fimicola CBS 482.64T as an out-group. Bar, 0.02 substitutions per nucleotide position.
4. Discussion
Strains KNUF-21-020 and KNUF-21-021 were collected from soil in Chungnam province, South Korea, and identified as Triangularia manubriata sp. nov. To the best of our knowledge, this is the first report of the genus Triangularia in Korea. The genus Triangularia was originally classified under the family Lasiosphaeriaceae [Citation2] in the early nineteenth century as a fungal species growing on a decayed substrate that produced ellipsoidal or cylindrical melanized ascospores [Citation15–18]. However, based on molecular analyses, the defined genera belonging to Lasiosphaeriaceae were revealed as polyphyletic [Citation19–21]. Thus, the family Podosporaceae was reclassified from Lasiosphaeriaceae by Wang et al. and established with three genera, namely, Cladorrhinum, Podospora, and Triangularia, based on phylogenetic analysis [Citation2]. At the same time, various species in several genera that previously belonged to Lasiosphaeriaceae, including Sordaria, Apiosordaria, Cercospora, and Zopfiella were redefined to the genus Triangularia, and several species in the genus Triangularia were reclassified to other genera [Citation2–4]. Thus, Triangularia is phylogenetically complex, and our report of T. manubriata sp. nov. is expected to contribute to the resolution of this phylogeny [Citation4].
Some species in this genus produce different secondary metabolites [Citation5]. For example, T. anserina produces larvicidal bioactive compounds called anthraquinones and the antibacterial and antifungal compound 1,4-benzoquinone [Citation22]. Antineoplastic and inflammatory metabolites of polyketide synthase and nonribosomal peptide synthetase hybrids have been reported from T. effusa [Citation23]. Additionally, T. bambusae, a species closely related to strain KNUF-21-020, was reported to produce the antifungal metabolite tetrahydrofuran [Citation24]. Therefore, further research on the secondary metabolites of KNUF-21-020 and KNUF-21-021 is necessary to determine the potential application of these strains.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Frąc M, Hannula SE, Bełka M, et al. Fungal biodiversity and their role in soil health. Front Microbiol. 2018;9:707. doi: 10.3389/fmicb.2018.00707.
- Wang XW, Bai FY, Bensch K, et al. Phylogenetic re-evaluation of Thielavia with the introduction of a new family Podosporaceae. Stud Mycol. 2019;93(1):155–252. doi: 10.1016/j.simyco.2019.08.002.
- Huang SK, Hyde KD, Mapook A, et al. Taxonomic studies of some often over-looked Diaporthomycetidae and Sordariomycetidae. Fungal Divers. 2021;111(1):443–572. doi: 10.1007/s13225-021-00488-4.
- Marin-Felix Y, Miller AN, Cano-Lira JF, et al. Re-evaluation of the order Sordariales: delimitation of Lasiosphaeriaceae s. str., and introduction of the new families Diplogelasinosporaceae, Naviculisporaceae, and Schizotheciaceae. Microorganisms. 2020;8(9):1430. doi: 10.3390/microorganisms8091430.
- Charria-Girón E, Surup F, Marin-Felix Y. Diversity of biologically active secondary metabolites in the ascomycete order Sordariales. Mycol Prog. 2022;21:1–33.
- Das K, You YH, Lee SY, et al. A new species of Thelonectria and a new record of Cephalotrichum hinnuleum from Gunwi and Ulleungdo in Korea. Mycobiology. 2020;48(5):341–350. doi: 10.1080/12298093.2020.1807454.
- White T, Bruns T, Lee S, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, et al., editors. PCR protocols: a guide to methods and applications. San Diego: Academic Press; 1990. p. 315–322.
- Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol. 1990;172(8):4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990.
- Donaldson GC, Ball LA, Axelrood PE, et al. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol. 1995;61(4):1331–1340. doi: 10.1128/aem.61.4.1331-1340.1995.
- O'Donnell K, Cigelnik E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol Phylogenet Evol. 1997;7(1):103–116. doi: 10.1006/mpev.1996.0376.
- Larkin MA, Blackshields G, Brown NP, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404.
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425.
- Kumar S, Stecher G, Li M, et al. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096.
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16(2):111–120. doi: 10.1007/BF01731581.
- Lundqvist N. Nordic Sordariaceae s. lat. Symb Bot Ups. 1972;20:1–374.
- Hilber O, Hilber R. Einige Anmerkungen zu der Gattung Cercophora Fuckel (Lasiosphaeriaceae). Zeitschrift Für Mykol. 1979;45:209–233.
- Guarro J. A synopsis of the genus Zopfiella (Ascomycetes, Lasiosphaeriaceae). Syst Ascomycet. 1991;10:79–112.
- Bell A. Podospora petrogale fungi: Sordariales: Lasiosphaeriaceae, a new species from Australia. Mueller. 2000;12:235–240.
- García D, Stchigel AM, Cano J, et al. A synopsis and re-circumscription of Neurospora (syn. Gelasinospora) based on ultrastructural and 28S rDNA sequence data. Mycol Res. 2004;108(10):1119–1142. doi: 10.1017/s0953756204000218.
- Cai L, Jeewon R, Hyde KD. Molecular systematics of Zopfiella and allied genera: evidence from multi-gene sequence analyses. Mycol Res. 2006;110(4):359–368. doi: 10.1016/j.mycres.2006.01.007.
- Huhndorf SM, Miller AN, Fernández FA. Molecular systematics of the Sordariales: the order and the family Lasiosphaeriaceae redefined. Mycologia. 2017;96(2):368–387. doi: 10.1080/15572536.2005.11832982.
- Matasyoh JC, Dittrich B, Schueffler A, et al. Larvicidal activity of metabolites from the endophytic Podospora sp. against the malaria vector Anopheles gambiae. Parasitol Res. 2011;108(3):561–566. doi: 10.1007/s00436-010-2098-1.
- Takahashi S, Kagasaki T, Furuya K, et al. Apiodionen, inhibitor of topoisomerase and suppressor of chemiluminescence: taxonomy, fermentation, isolation, structural elucidation and biological activity. Annu Rep Sankyo Res Lab. 1992;44:119–127.
- Nakagawa F, Kodama K, Furuya K, et al. New strains of botryodiplodin-producing fungi. Agric Biol Chem. 1979;43(7):1597–1598. doi: 10.1080/00021369.1979.10863669.

