Abstract
There is increasing evidence that holocarpic oomycetes, i.e., those converting their entire vegetative thallus into zoospores upon maturation, are a phylogenetically diverse group in both freshwater and marine ecosystems. Most of the known holocarpic oomycete species diverge before the main split of Peronosporomycetes and Saprolegniomycetes and are, thus, termed as early-diverging oomycetes. In environmental sequencing studies, it was revealed that of the early-diverging genera especially Sirolpidium, Miracula, and Diatomophthora are widespread. As in these studies especially the Arctic Ocean seemed to harbor many undiscovered species, sampling was conducted at the Blávík research station on Fáskrúðsfjörður in the East Fjords of Iceland, where there is both an influence from the Arctic Ocean and the North Atlantic. During the screening for infected diatoms, a parasitoid was found in the marine diatom genus Melosira, which is one of the most abundant genera in arctic ecosystems. Molecular phylogenetics and morphological characterization revealed that the parasitoid belonged to the genus Miracula and corresponded to one of the lineages previously found in single-cell sequencing. Thus, the current study both contributes to the knowledge of the genus Miracula and the increasing diversity of the genus suggests that the many linages found in environmental sequencing which can still not be associated with known species might represent actual species to be discovered in future studies.
1. Introduction
Diatoms are responsible for a major share of the global photosynthesis, especially in marine ecosystems [Citation1,Citation2]. At the same time, they are attacked by various viruses [Citation3,Citation4], bacteria [Citation5,Citation6], fungi [Citation7,Citation8], and oomycetes [Citation9–12]. One of the most widespread oomycete genera affecting diatoms is the recently established genus Miracula [Citation13], which has been isolated both from marine [Citation14,Citation15] and freshwater [Citation16,Citation17] environments, while environmental sequencing studies also suggested its widespread presence in the Arctic Ocean [Citation18]. The genus Miracula belongs to the early-diverging oomycetes [Citation19], and so far contains exclusively holocarpic parasitoids of diatoms. Despite the richness and abundance of potential diatom hosts in the Arctic [Citation20], arctic ecosystems are so far hardly explored in terms of phytoplankton pathogens. In some of our previous studies, we already found several holocarpic oomycete diatom parasitoids in Iceland, including three new species of the genus Miracula, namely Miracula islandica [Citation14], M. einbuarlaekurica [Citation17], and M. blauvikensis [Citation15], as well as one new species of Lagena, Lagena ausuennarstadhirensis [Citation21]. The recently found species, M. blauvikensis was the first species of the genus that was found to be associated with a larger cluster of sequences obtained from amplified genomes of isolated single cells [Citation11,Citation15]. This finding prompted the search for additional species of the genus and it was the aim of this study to screen arctic coastal diatoms for the presence of additional parasitoids of the genus Miracula and to characterize them.
2. Materials and methods
2.1. Oomycete screening, sampling, and morphological characterization
Samples with coastal diatoms were obtained at the Blávík Biological Arctic Research Station, located on Fáskrúðsfjörður in the East Fjords of Iceland, by scraping loose biofilm with mineralic material (sand and seashell fragments) from flat stones in rockpools and by collecting plankton as described earlier [Citation15,Citation22,Citation23] on August 6 and 8 2022 and April 4 2023. Samples were screened in 12.5 cm Petri dishes using an inverted microscope (Eclipse T1000, Nikon, Tokyo, Japan). Infected diatoms were isolated using a micropipette (Transferpette 10 µl, Brandt, Wertheim, Germany) as described earlier [Citation12]. The infected hosts were collected into both ethanol (for obtaining specimens for deposit) and water (for DNA extraction).
A low percentage of cells of the centric filamentous diatom Melosira sp. was found to be infected by a holocarpic oomycete, but infections increased after incubation for several days as described previously [Citation17]. The morphology and life-cycle stages of the parasitoid were documented using a compound microscope equipped with DIC (Imager M2, Zeiss, Oberkochen, Germany). Measurements were done using AxioVision v7 (Zeiss, Oberkochen, Germany), using calibrated photographs obtained by an AxioCam (Zeiss, Oberkochen, Germany). Dual cultures were attempted as outlined in [Citation15]. While those attempts were unsuccessful in 2022, a dual culture could be achieved in 2023 and host and parasitoid dual cultures are available from the authors upon request.
2.2. PCR, sequencing, and phylogenetics
PCR and sequencing for obtaining partial nuclear small ribosomal subunit (18S) and mitochondrial cytochrome oxidase subunit 2 (cox2) sequences was done as described previously [Citation23]. While the 18S locus has been widely used for phylogenetic investigations in early-diverging oomycetes (e.g., [Citation13]), the cox2 molecular barcode is especially suited for species-level differentiation in oomycetes [Citation24]. Sequences were deposited in GenBank under the accession numbers PP492446 (isolate from 2022) and PP492447 (isolate from 2023) for SSU and PP503200 (isolate from 2022) and PP503201 (isolate from 2023) for cox2.
Alignments were done using muscle [Citation25], as implemented in MEGA11 [26] using −600 as gap opening penalty and −8 as gap extension penalty. The alignments are provided as Supplementary File 1 (18S) and Supplementary File 2 (cox2). Phylogenetic analyses were done on the TrEase webserver [Citation26] using FastTree2 [Citation27] for minimum evolution (ME) and RAxML [Citation28] for maximum-likelihood (ML) inference, each with the standard settings of the server. Phylogenetic trees were visualized and sorted stepwise up in MEGA11 [Citation29], and exported as encapsulated metafile for adjusting and editing labels in Microsoft PowerPoint 2013 (Microsoft, Redmond, CA).
3. Results
3.1. Morphology and life-cycle
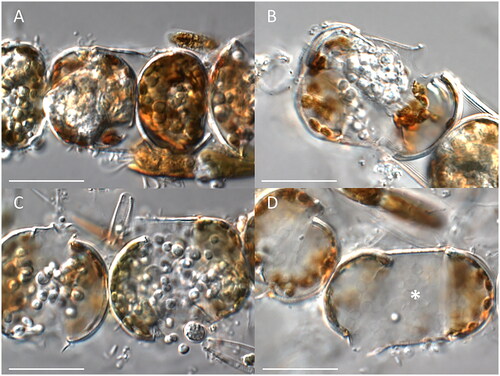
The infection cycle started when zoospores settled and encysted on the surface of the host frustules. Subsequently, the cysts germinated to produce a very fine penetration tube extending through the girdle bands into the host cytoplasm. In early stages, the thallus was barely visible due to the dense plastid arrangement in the host. Once established, the thallus underwent rapid growth and became more visible (). In contrast to most other holocarpic oomycetes parasitizing diatoms, the host phaeoplasts retained their integrity over most of the maturation process of the parasitoid and also the cytoplasm did not show intense degradation throughout the longer part of the infection. In some cases, phaeoplasts seemed little affected even after spore discharge (), while sometimes the cytoplasm and phaeoplasts degraded (e.g., ). Host cells normally contained one or two spherical to irregular-shaped endobiotic holocarpic thalli, but occasionally more than 5, compacted tightly within the frustule. The opaque silvery amorphous thalli were 15–50 μm by 20–70 μm in diameter when singly, but smaller in multiple infections. At thallus maturity, the thalli were swelling, forcing apart the host valves until the thalli, which very surrounded by a very thin, colorless wall, came in contact with the surrounding medium. At this stage of the infection, well-differentiated zoospores were visible, which usually encysted before vigorous swimming. While the thalli became exposed to the surrounding medium, usually 1–2 short, broad conical discharge tubes developed. Subsequently, the cysts, which were 5–8 µm in diameter and surrounded by a very thin cell wall, germinated by producing a single zoospore. Cyst germination was mostly almost simultaneous, and the zoospores produced were broadly reniform to sublenticular 6–8 µm long, 5–7 µm broad, swimming away randomly in uneven rotation after leaving through the exit tubes. After few minutes of motility, zoospores started to rest. Resting cell germination was not observed and no sexual stages could be differentiated.
Figure 1. Morphological characteristics of Miracula polaris. (A) Early infection stages until zoospore maturation; (B, C) late stages of infection: (B) formation of primary cysts; (C) zoospore release; (D) empty cysts (asterisk) in the host cell after the release of secondary zoospores. The scale bar equals 50 µm in all pictures.
3.2. Phylogeny
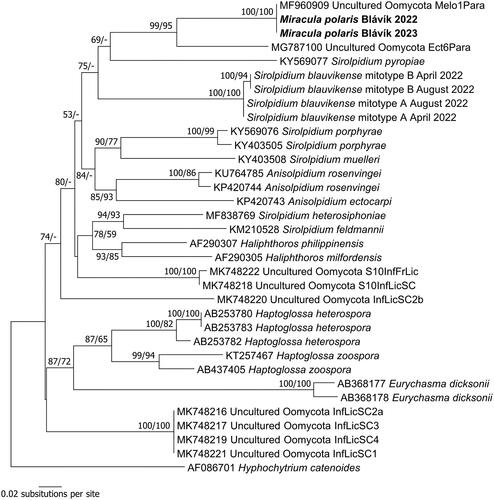
In nrSSU sequences, the parasitoids from Melosira sp. from 2022 and 2023 were identical and grouped together with maximum support in the ME analysis (). The sequences were found to be almost identical to a sequence obtained from a single cell of Melosira sp. in a previous study [Citation11], with just one indel in the aligned nrSSU sequence. The monophyly of this group received high bootstrap support in the ME phylogenetic inference but no support in the ML analysis. This group was resolved as the sister clade to Miracula islandica parasitizing Minidiscus, but with low support. The two species grouped together with a clade consisting of M. helgolandica parasitizing Pseudo-nitzschia pungens and two lineages known only from environmental sequencing. This clade with three parasitoids that have been morphologically characterized was resolved as monophyletic with maximum support in both ME and ML. Its sister clade was the freshwater clade of Miracula with M. einbuarlaekurica and M. moenusica, which received strong bootstrap support for its monophyly (92% in ME and 96% in ML). The two clades grouped together with maximum support in both ME and ML. Together with a deeply divergent clade that contained M. blauvikensis, they represented the genus Miracula, which monophyly received maximum support in both ME and ML. Interestingly, some higher-level relationships were resolved in the ME-based tree, e.g., the monogeneric Pontismatales were resolved as monophyletic with moderate support (81%) in the ME inference, and the grouping of the filamentous oomycete lineages (the crown clades plus Haliphthorales) received weak to moderate bootstrap support in ME (70%).
Figure 2. Phylogenetic reconstruction in minimum evolution based on partial nrSSU sequences. Numbers near nodes represent bootstrap support values in minimum evolution and maximum likelihood. A minus sign denotes lack of support for the presented or an alternate topology.
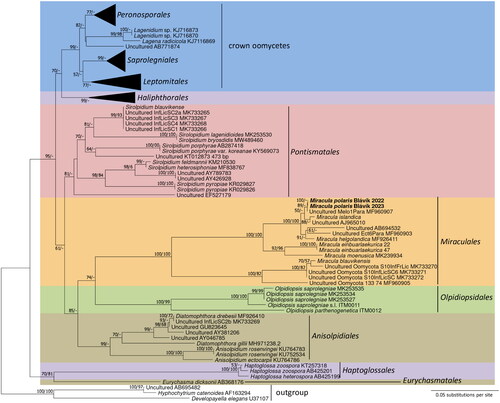
In the phylogeny based on cox2 sequences (), the parasitoid on Melosira sp. found in this study revealed to be identical to the single-cell sample from the same host genus by Garvetto et al. [Citation11]. Thus, this group that also had a long branch until the next bifurcation received maximum support in both ME and ML. It was further grouped together with a sequence from another single-cell sample. Similar to the situation found in previous studies (e.g., [Citation23]), cox2 did not provide good resolution above the species level, due to its high variability.
4. Taxonomy
Based on its phylogenetic position, host, and morphological characteristics, the parasitoid of Melosira sp. found in this study is described as a new species.
Miracula polaris Buaya & Thines, sp. nov., MycoBank MB 852870
Etymology – Referring to the habitat in close to the Arctic Circle.
Description – Thallus parasitic in the diatom genus Melosira, silvery, 15–50 μm by 20–70 μm in diameter when singly, smaller in multiple infections, surrounded by a very thin, colorless wall at maturity, leading to a swelling of infected cells, forcing apart the host valves. Discharge tubes broad, conical, 10–15 µm at the base and 5–8 µm at the top. Primary zoospores roundish, 5–8 µm in diameter, encysting within the host cell before release. Cyst germination often almost simultaneous. Secondary zoospores broadly reniform to sublenticular 6–8 µm long, 5–7 µm broad, swimming away randomly in uneven rotation after leaving through the exit tubes. Sexual reproduction not observed.
Diagnosis – Differs from known species of Miracula by producing cysts inside the thallus before escaping from discharge tubes.
Type: Iceland, Fáskrúðsfjörður, Blávík, parasitic in cells of Melosira sp., collected by Marco Thines on April 4 2023, deposited in the Herbarium Senckenbergianum (FR) under the accession number FR-0046168.
5. Discussion
Holocarpic oomycetes are widespread yet elusive organisms, due to their small size and their mostly obligate biotrophic nature [Citation19]. However, as their importance for natural ecosystems is increasingly realized [Citation30], research in these organisms has accelerated over the past years [Citation10,Citation13,Citation22,Citation23,Citation31,Citation32]. With the broader use of environmental barcoding, it became apparent that oomycetes are potentially diverse in the marine realm [Citation11,Citation18,Citation33].
The study of Hassett et al. [Citation18] suggested that the Arctic Ocean harbors a huge diversity of holocarpic oomycetes and Garvetto et al. [Citation11] provided host information for some lineages that enabled a more targeted screening for oomycetes in marine diatoms. An interesting finding of Garvetto et al. [Citation11] was that the genus Melosira, which is widespread throughout polar regions and can form massive blooms beneath sea ice [Citation34], is host to a species of Miracula. In this study, we were able to isolate and characterize the parasitoid and to establish a dual culture together with its host. Notably, the cox2 sequence of Miracula polaris was identical to the single-cell sample used by Garvetto et al. [Citation11], confirming the identity of the organisms and highlighting that M. polaris might be a widespread species.
As still most lineages found by Hassett et al. [Citation18] and Garvetto et al. [Citation11] have not been isolated, it seems likely that many more species await discovery in Polar Regions and beyond. At the same time, the abundance of holocarpic oomycetes in environmental sequencing studies provides hints that oomycete parasitoids might be a major force regulating diatom blooms in the global oceans [Citation35].
Author contributions
AB and MT conceived the study. MT conducted the sampling. AB conducted laboratory experiments, including pathogen isolation, microscopic observations, PCR, and sequencing. MT did the phylogenetic analyses. MT wrote the manuscript with contributions from AB.
Alignment_nrSSU.docx
Download MS Word (20.5 KB)Alignment_cox2.docx
Download MS Word (16.2 KB)Acknowledgements
This study was enabled by a stay at the Blávík Arctic Research Station on Fáskrúðsfjörður, Fjarðabyggð, Iceland.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Miettinen A. Diatoms in Arctic regions: potential tools to decipher environmental changes. Polar Biol. 2018;18:220–226. doi: 10.1016/j.polar.2018.04.001.
- Gilbertson R, Langan E, Mock T. Diatoms and their microbiomes in complex and changing polar oceans. Front Microbiol. 2022;13:786764.
- Nagasaki K, Tomaru Y, Takao Y, et al. Previously unknown virus infects marine diatom. Appl Environ Microbiol. 2005;71(7):3528–3535. doi: 10.1128/AEM.71.7.3528-3535.2005.
- Toyoda K, Kimura K, Osada K, et al. Novel marine diatom ssRNA virus NitRevRNAV infecting Nitzschia reversa. Plant Ecol Evol. 2019;152(2):178–187. doi: 10.5091/plecevo.2019.1615.
- Amin SA, Parker MS, Armbrust EV. Interactions between diatoms and bacteria. Microbiol Mol Biol Rev. 2012;76(3):667–684. doi: 10.1128/MMBR.00007-12.
- Van Tol HM, Amin SA, Armbrust E. Ubiquitous marine bacterium inhibits diatom cell division. ISME J. 2017;11(1):31–42. doi: 10.1038/ismej.2016.112.
- Scholz B, Küpper FC, Vyverman W, et al. Effects of eukaryotic pathogens (Chytridiomycota and Oomycota) on marine benthic diatom communities in the Solthörn tidal flat Southern North Sea, Germany. Eur J Phycol. 2016;51(3):253–269. doi: 10.1080/09670262.2015.1134814.
- Chambouvet A, Monier A, Maguire F, et al. Intracellular infection of diverse diatoms by an evolutionary distinct relative of the fungi. Curr Biol. 2019;29(23):4093–4101.e4. doi: 10.1016/j.cub.2019.09.074.
- Scholz B, Küpper FC, Vyverman W, et al. Eukaryotic pathogens (Chytridiomycota and Oomycota) infecting marine microphytobenthic diatoms – a methodological comparison. J Phycol. 2014;50(6):1009–1019. doi: 10.1111/jpy.12230.
- Thines M, Nam B, Nigrelli L, et al. The diatom parasite Lagenisma coscinodisci (Lagenismatales, Oomycota) is an early diverging lineage of the Saprolegniomycetes. Mycol Prog. 2015;14(75):1–7.
- Garvetto A, Nezan E, Badis Y, et al. Novel widespread marine oomycetes parasitising diatoms, including the toxic genus Pseudo-nitzschia: genetic, morphological, and ecological characterisation. Front Microbiol. 2018;9:2918.
- Buaya AT, Ploch S, Kraberg A, et al. Phylogeny and cultivation of the holocarpic oomycete Diatomophthora perforans comb. nov., an endoparasitoid of marine diatoms. Mycol Prog. 2020;19(5):441–454. doi: 10.1007/s11557-020-01569-5.
- Buaya AT, Ploch S, Hanic L, et al. Phylogeny of Miracula helgolandica gen. et sp. nov. and Olpidiopsis drebesii sp. nov., two basal oomycete parasitoids of marine diatoms, with notes on the taxonomy of Ectrogella-like species. Mycol Prog. 2017;16(11–12):1041–1050. doi: 10.1007/s11557-017-1345-6.
- Buaya AT, Scholz B, Thines M. A new marine species of Miracula (Oomycota) parasitic to Minidiscus sp. in Iceland. Mycobiology. 2021;49(4):355–362. doi: 10.1080/12298093.2021.1952813.
- Buaya A, Thines M. Miracula blauvikensis: a new species of Miracula from Iceland, and report of a co-cultivation system for studying oomycete–diatom interactions. Fungal Syst Evol. 2022;10(1):169–175. doi: 10.3114/fuse.2022.10.07.
- Buaya AT, Thines M. Miracula moenusica, a new member of the holocarpic parasitoid genus from the invasive freshwater diatom Pleurosira laevis. Fungal Syst Evol. 2019;3(1):35–40. doi: 10.3114/fuse.2019.03.04.
- Buaya AT, Thines M. Miracula einbuarlaekurica sp. nov., a new holocarpic endoparasitoid species from pennate freshwater diatoms in Iceland. Mycology. 2022;13(2):153–161. doi: 10.1080/21501203.2021.1980446.
- Hassett BT, Thines M, Buaya A, et al. A glimpse into the biogeography, seasonality, and ecological functions of arctic marine Oomycota. IMA Fungus. 2019;10(1):6. doi: 10.1186/s43008-019-0006-6.
- Beakes GW, Thines M. Hyphochytriomycota and Oomycota. In: Archibald JM, Simpson AGB, Slamovits CH, editors. Handbook of the protists. Heidelberg: Springer Verlag; 2017. p. 435–505.
- Stoof‐Leichsenring KR, Pestryakova LA, Epp LS, et al. Phylogenetic diversity and environment form assembly rules for Arctic diatom genera—a study on recent and ancient sedimentary DNA. J Biogeogr. 2020;47(5):1166–1179. doi: 10.1111/jbi.13786.
- Thines M, Buaya AT. Lagena—an overlooked oomycete genus with a wide range of hosts. Mycol Prog. 2022;21(8):66.
- Buaya A, Tsai I, Thines M. Pontisma blauvikense sp. nov. the first member of the early‐diverging oomycete genus Pontisma parasitizing brown algae. J Eukaryot Microbiol. 2023;70(2):e12957. doi: 10.1111/jeu.12957.
- Buaya AT, Tsai I, Klochkova TA, et al. Introducing a new pathosystem for marine pathogens – the green alga Urospora neglecta and its pathogen Sirolpidium litorale sp. nov. Mycol Prog. 2023b;22(86):1–7.
- Choi YJ, Beakes G, Glockling S, et al. Towards a universal barcode of oomycetes – a comparison of the cox1 and cox2 loci. Mol Ecol Resour. 2015;15(6):1275–1288. doi: 10.1111/1755-0998.12398.
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340.
- Mishra B, Ploch S, Weiland C, et al. The TrEase web service: inferring phylogenetic trees with ease. Mycol Prog. 2023;22(84):1–6.
- Price MN, Dehal PS, Arkin AP. Arkin AP FastTree 2 – approximately maximum-likelihood trees for large alignments. PLOS One. 2010;5(3):e9490. doi: 10.1371/journal.pone.0009490.
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033.
- Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 2021;38(7):3022–3027. doi: 10.1093/molbev/msab120.
- Scholz B, Guillou L, Marano AV, et al. Zoosporic parasites infecting marine diatoms – a black box that needs to be opened. Fungal Ecol. 2016;19:59–76. doi: 10.1016/j.funeco.2015.09.002.
- Gachon CM, Strittmatter M, Badis Y, et al. Pathogens of brown algae: culture studies of Anisolpidium ectocarpii and A. rosenvingei reveal that the Anisolpidiales are uniflagellated oomycetes. Eur J Phycol. 2017;52(2):133–148. doi: 10.1080/09670262.2016.1252857.
- Badis Y, Klochkova TA, Brakel J, et al. Hidden diversity in the oomycete genus Olpidiopsis is a potential hazard to red algal cultivation and conservation worldwide. Eur J Phycol. 2020;55(2):162–171. doi: 10.1080/09670262.2019.1664769.
- Langer JA, Sharma R, Nam B, et al. Cox2 community barcoding at Prince Edward Island reveals long-distance dispersal of a downy mildew species and potentially marine members of the Saprolegniaceae. Mycol Progress. 2021;20(4):509–516. doi: 10.1007/s11557-021-01687-8.
- Poulin M, Underwood GJ, Michel C. Sub-ice colonial Melosira arctica in Arctic first-year ice. Diatom Res. 2014;29(2):213–221. doi: 10.1080/0269249X.2013.877085.
- Metfies K, Hessel J, Klenk R, et al. Uncovering the intricacies of microbial community dynamics at Helgoland Roads at the end of a spring bloom using automated sampling and 18S meta-barcoding. PLOS One. 2020;15(6):e0233921. doi: 10.1371/journal.pone.0233921.