ABSTRACT
Pancreatic islet β-cells weaken under oxidative stress. In this study, human pancreatic islet-derived 1.1B4 cells were exposed to H2O2 and analysed using a human microarray, which revealed that heme oxygenase 1 (HMOX1), glutamate-cysteine ligase, early growth response 1, nuclear receptor subfamily 4 group A member 3 (NR4A3) and jun B proto-oncogene were upregulated, whereas superoxide dismutase 1 and catalase were not. Expression of NR4A3 rapidly increased after H2O2 addition, and the 1.1B4 cells treated with siRNA targeting NR4A3 became sensitive to H2O2; further, HMOX1 expression was strongly inhibited, suggesting that NR4A3 is an oxidative stress-responsive transcription factor that functions through HMOX1 expression in pancreatic islet β-cells. Expression of cyclin E1 and cyclin-dependent kinase 1 was also inhibited by siRNAs targeting NR4A3.
Introduction
Methods for restoring the decline in β-cells frequently observed in type 2 diabetes are needed [Citation1]. Elevated glucose concentrations have been suggested to increase the levels of reactive oxygen species (ROS) in β-cells [Citation2]. Chronically high ROS levels can decrease insulin gene expression and accelerate apoptosis [Citation2, Citation3]. Pancreatic β-cells are more sensitive to oxidative stress than other cells because these express antioxidant enzymes at a lower level than other cells [Citation2]. However, the effect of oxidative stress on β-cells is not well understood [Citation4].
High oxidative stress induces cell damage in several cell types. The regulatory mechanisms by which H2O2 modulates transcription factors are summarised in a review [Citation5], which reported that many sensing proteins and oxidants regulate transcription factors to induce apoptosis. Oxidative stress also induces the expression of heme oxygenase-1 (HMOX1) that acts as an antioxidant defence system [Citation6].
We previously found that oxidative stress induced by H2O2 increased the expression of the nuclear receptor subfamily 4, Group A member 1 (NR4A1) and NR4A3 genes in HUC-F2 fibroblasts, and that inhibition of NR4A1 with specific siRNA triggered an increase in apoptosis following the addition of H2O2 [Citation7]. The NR4A orphan nuclear receptor family of transcription factors is rapidly and strongly upregulated following exposure to stressful stimuli [Citation8]. Lee et al. [Citation9] and Volakakis et al. [Citation10] reported that expression of NR4A genes is modulated by oxidative stress.
In this study, we aimed to analyse gene expression in β-cells in response to ROS using H2O2 and human pancreatic β-cell-derived hybrid cells (1.1B4 cells). We analysed the effect of H2O2 on the expression of genes in 1.1B4 cells using a human microarray. We also analysed the expression of genes in NR4A3 siRNA-treated cells to determine the effect of decreased NR4A3 mRNA on the expression of genes in 1.1B4 cells.
Materials and methods
Microarray and software
Whole Human Genome Oligo Microarray Kit 4 × 44 K ver. 2.0, Feature Extraction Software 9.5.1.1 (Agilent Technologies, Palo Alto, CA, USA), GeneSpring GX10 v 7.3.1 (Agilent Technologies), and Spotfire software (TIBCO, NTTCom, Tokyo, Japan) were used.
Biological Resources: human pancreatic islet-derived cells (1.1B4 cells)
Cells of 1.1B4 (ECACC No. 10012801) [Citation11], a hybrid of normal human islet cells and transformed PANC-1 cells, were obtained from DS Pharma Biomedical Co. Ltd., Osaka, Japan.
Cell culture of 1.1B4 cells and RT–qPCR
The 1.1B4 cells were cultured in RPMI-1640 medium. Cell viability was analysed using an MTT assay [Citation12]. Total RNA was extracted using Isogen [Citation13] or RNeasy Mini Kit. The expression was measured using RT-qPCR. The procedures are described in detail in the Supplemental Material and Supplemental Table S1.
RNA interference of NR4A3
NR4A3 Silencer Select siRNA was used according to the ABI protocol, with Silencer Select Negative Control #1 siRNA and nuclease-free water (NFW) (ABI) as negative controls. The efficiency was measured by RT-qPCR. The procedures are described in detail in the Supplemental Material.
Analysis of gene expression in 1.1B4 cells by microarray
Experiment A: H2O2 treatment
B4 cells (n = 12) were seeded at 1.5 × 105 cells/ml in the microplate (Agilent Technologies), and 24 h later, six wells were incubated with 100 µM H2O2 for 4 h (H2O2 treated group). Total RNA was extracted from the 12 wells using the RNeasy Mini Kit according to the manufacturer’s instructions. The quality of the 12 RNA samples was assessed, and an aliquot of three RNA samples from the same group was combined into one microarray sample, resulting in a total of two microarray samples for the H2O2 treated group and two for the control group. The four samples were analysed using a human microarray according to the manufacturer’s protocol. After reading, one control microarray was scratched; the expression was compared to the average of two treated array samples and one control. The 12 RNA samples were also used individually for RT-qPCR. (See supplemental Fig. S7)
Experiment B: siNR4A3 treatment
The 1.1B4 cells (n = 22) were incubated for 24 h. Half (n = 11) were treated with siNR4A3, and the other half (n = 11) were treated with NFW. Forty-eight hours later, cells in two wells from each group were treated with 100 µM H2O2 for 2 h. After 24 h, mRNA was purified. Aliquots of three RNA samples without H2O2 were combined into one microarray sample (three siRNA-treated and three control) and subjected to microarray analysis, as described for experiment A. Aliquots of two RNA samples from the H2O2-treated group were also combined and analysed (n = 2, siRNA-treated H2O2-treated sample and siRNA-treated without H2O2 treatment). For RT-qPCR, 22 RNA samples were used individually. (See supplemental Fig. S7)
Statistical analysis
Student’s t-test, ANOVA with Tukey’s HSD post hoc test, and pairwise t-test (R Core Team; R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria; 2017 URL https://www.R-project.org/) were used.
Results
Effect of H2O2 on 1.1B4 cell growth and gene expression analysed by DNA microarray
Human 1.1B4 cells were exposed to 0, 10, 20, 30, 40, 50, 100 or 200 µM H2O2, and cell viability was measured using the MTT assay (Supplemental Fig. S1). We observed no toxicity in response to H2O2 up to 50 µM; growth was limited at 100 µM H2O2 and was completely suppressed at 200 µM. Therefore, to investigate the antioxidant system, 1.1B4 cells were treated with 100 µM H2O2 and analysed using a human microarray. The results were submitted to the Gene Expression Omnibus (GEO) database of NCBI (accession number GSE83369). (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE83369). A total of 2903 genes were upregulated by more than two-fold in untreated cells, and 2283 genes were downregulated by less than half. Most genes were classified as unknown or others, and genes classified as nucleotide metabolism-related comprised the major group (, Supplemental Fig. S2, S3). The expression of several genes was confirmed using RT-qPCR (, ). The expression ratio of the antioxidant enzyme HMOX1 ((b)) was high and upregulated; however, those of SOD1 and CAT ((a)) remained unchanged, and their relative expression was considerably lower than that of HMOX1. In contrast, the levels of peroxide-reducing GPX1 and the antioxidant enzyme GCLC were increased, but their expression ratio was low. Among the antioxidant pathways [Citation5,Citation6], the levels of KEAP1 ((d)), CREB, HSF, HIF1, TP53, NF-κB and SP1 were unchanged. Among the transcription factors upregulated by H2O2 (Marinho et al. [Citation5]), FOSB ((c)), and JUNB were strongly upregulated, suggesting that the AP-1 pathway was upregulated. Insulin was downregulated, and the apoptosis marker caspases were upregulated. Many genes whose functions are unknown were upregulated or downregulated. Among these, NR4A3, an orphan receptor, was significantly upregulated ((e)). Considering this result, we measured the effect of H2O2 concentration on NR4A3 expression ((a)). The NR4A3 gene was significantly upregulated by H2O2 (used at a concentration of 100 µM for 4 h) ((b)). Interestingly, this expression rapidly increased and then decreased to its original levels by 24 h, suggesting that NR4A3 is an early responder to H2O2 stress. To further investigate the role of NR4A3, we treated 1.1B4 cells with siRNA targeting NR4A3 followed by H2O2 treatment, and then measured cell viability. The treatment of 1.1B4 cells with siRNA decreased cell viability, which was strongly inhibited by the addition of H2O2 after siRNA treatment ().
Figure 1. Effect of H2O2 addition on expression of various genes. A. Human 1.1B4 cells were treated with H2O2 as described in and Supplemental Material. For RT-qPCR analysis of CAT under Condition A described in the Supplemental Material, 12 RNA samples from the two groups obtained for microarray were used, and the value obtained was the expression ratio to GAPDH expression. (* indicates a significant difference of p < 0.05 by t-test). B-F. Levels of HMOX1, FOSB, KEAP1, NR4A3, and EGR1, respectively, were measured.

Figure 2. Effect of H2O2 concentration on NR4A3 gene expression and time course of expression. A. Human 1.1B4 cells (1.0 × 105 cells/ml) were incubated in 2 ml of culture medium, and 24 h later, the medium was changed to individual concentrations of H2O2-containing medium. After 4 as well as 24 h, total RNA was extracted using Isogen reagent. The expression of NR4A3 was measured by RT-qPCR under Condition B (n = 3) described in the Supplemental Material. B. After addition of 100 µM H2O2, the cells were incubated for various time periods and RNA was extracted (n = 3). ß-Actin was used as a control. (* p < 0.05, ** p < 0.01).

Figure 3. Effect of siRNA targeting NR4A3 on cell viability. A. After addition of siNR4A3 to 1.1B4 cells, total RNA was extracted and expression of NR4A3 was measured by RT-qPCR using the Cyber green method under Condition B described in the Supplemental Material (n = 3; * p < 0.05). B. After addition of siNR4A3, the 1.1B4 cells were incubated with 100 mM of H2O2, and cell viability was measured by MTT assay (n = 5). NFW: nuclease free water-added cell; siNR4A3: siRNA of NR4A3-treated cell.

Table 1. Comparison of the expression between H2O2-treated 1.1B4 cells and control cells using microarray and RT-qPCR.
Comprehensive gene expression analysis of 1.1B4 cells treated with siRNA targeting NR4A3 by DNA microarray
Next, we performed a comprehensive gene expression analysis of 1.1B4 cells using a human microarray after treatment with siRNA targeting NR4A3. The data were submitted to the GEO database of NCBI under the accession number GSE86924 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE86924). We found that 1,044 genes were significantly upregulated by more than 1.5-fold, and 859 genes were significantly downregulated by less than 0.67-fold (Supplemental Fig. S4A, B, Supplemental Table S2). Changes in the expression of antioxidant enzymes, transcription factors related to H2O2 addition [Citation6], and several well-known genes are listed in . Some of these results were confirmed by RT-qPCR (, , Supplemental Fig. S5). NR4A3 expression was downregulated ((a)), that of the orphan receptor NR4A1 was unchanged, whereas that of NR4A2 was upregulated. Among the genes encoding antioxidant enzymes, the expression of HMOX1 was decreased ((b)). However, the expression of GCLC was only 0.88 times that of the control, and the expression of SOD1 was not significantly changed. We observed interesting gene expression responses of the antioxidant enzyme GLRX ((c)); its expression increased in contrast to that of HMOX1. The expression of most of the redox-sensitive transcription factors was unchanged, except for that of FOSB and JUNB, which was unchanged after siRNA addition but increased after H2O2 addition, suggesting that the AP-1 pathway operates independently. The expression of these genes in siNR4A3-treated cells was very low after H2O2 addition (). Among the cell cycle regulatory genes, the expression of CCNE1 (cyclin E) and CDK2 ((d)) was markedly decreased, as was that of several cyclin-dependent kinases. Therefore, it can be suggested that NR4A3 has a negative effect on the mRNA levels of cell cycle genes, especially cyclin E1 and cyclin-dependent kinase 2, directly or indirectly, and may cause growth inhibition.
Figure 4. Changes in gene expression in response to NR4A3 siRNA treatment and H2O2 treatment measured by RT-qPCR. RT-qPCR conditions are described in and Supplemental Material. A. NR4A3. B. HMOX1. C. GLRX. D. CDK2. E. EGR1. Pairwise t tests were used for the statistical analysis of post-hoc tests between NFW and siNR4A3 (* p < 0.05).

Table 2. Changes in gene expression in 1.1B4 cells in response to NR4A3 siRNA treatment and H2O2 treatment, measured by microarray and RT-qPCR.
Discussion
Recently, Chaffey et al. [Citation14] reported that 1.1B4 cells may not retain the phenotype expected from normal human β-cells. Therefore, we searched for the homology of the NR4A3 probe sequence used in this study (A_23_P398566) using MegaBlast at NCBI, and found that the probe sequence only matched the human NR4A3 mRNA and Macaca fascicularis brain cDNA clone, but did not match the corresponding rodent sequence. Therefore, the data presented in this study can be considered to correspond to human data (Supplemental Fig. S8). Chaffey et al. [Citation14] reported that the extremely limited availability of human pancreatic β-cell lines amenable to in vitro manipulation presents a clear barrier to progress in diabetes research. Primary human islets represent the gold standard for the study of β-cell function, but are available only intermittently. Therefore, the advent of a hybrid cell line obtained by the fusion of PANC-1 with a primary human β-cell, and which retains certain critical features of both offers a unique opportunity to study the development of persistent β-cell and diabetes research.
Because of the weak expression of antioxidant enzymes in islet β-cells [Citation2], these cells develop diabetes mellitus in response to oxidative stress. However, in other cells, H2O2 induced SOD1, SOD2, and CAT expression [Citation15]. In this study, HMOX and GCLC were strongly upregulated in H2O2-treated 1.1B4 cells (), whereas other major antioxidant enzymes were not upregulated. Therefore, HMOX1, GCLC, and the transcription factor NR4A3 are suggested to be responsible for the antioxidant defence system in pancreatic cells, and for defending against oxidative stress in diabetes, whereas CAT and GPX1 do not seem to be effective in these cells.
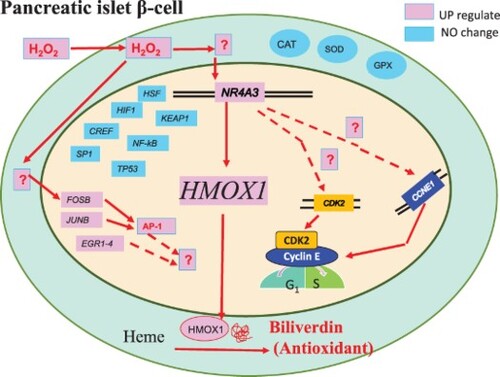
Alam and Cook [Citation6] have described four pathways involved in HMOX1 regulation. However, we demonstrated that the downregulation of HMOX1 by NR4A3 siRNA causes a loss of antioxidative resistance. It is suggested that NR4A3 is a fifth oxidative stress-responsive transcription factor for HMOX1 and may be a major one in β-cells (Graphical Abstract). The FOSB, JUNB, AP-1, and EGR1 genes were also upregulated ( and , (c,f)), suggesting that these factors might also play an important role; however, this requires further investigation.
The NR4A protein binds to NBRE (taggtca), or forms heterodimers and then binds to NuRE (aat(g/a)(c/t)ca) [Citation16]. The antisense sequences tgaccta and tgacatt were observed to be located upstream of HMOX1 (Supplemental Fig. S6A). CDK2 and CCNE1 also contained these sequences (Supplementary Fig. S6B and C), suggesting that these transcription factors are regulated by NR4A3 following H2O2 addition.
Robertson [Citation17] reported that the unrelenting deterioration of β-cells is related to chronic oxidative stress. In this study, insulin expression in 1.1B4 cells decreased by 0.02-fold after H2O2 addition as compared to that in control cells (), and diabetes led to increased oxidative stress, suggesting that β-cells decreased insulin production in diabetes. This suggests that drug discovery trials should include antioxidant protection of β-cells. However, antioxidant drugs are not currently clinically used for the treatment of human diabetes. We recently identified a strong antioxidant zeylaniin A [Citation18] in food products, which may be useful for the treatment of human diabetes in future. The limit of this study is the cells used is hybrid of normal human cells and transformed cells, but it is concluded that the results of this study show that HMOX1, GCLC, and the transcription factor NR4A3 are suggested to be responsible for the antioxidant defence system in pancreatic cells, and for defending against oxidative stress in human diabetes.
Geolocation information
Department of Life Science, Graduate School, Ochanomizu University, Bunkyo-ku, Tokyo, 112-8610, Japan.
Disclosure of interest
The authors report there are no competing interests to declare.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The datasets are available from the corresponding author upon reasonable request.
Additional information
Funding
References
- Tessem JS, Moss LG, Chao LC, et al. Nkx6.1 regulates islet β-cell proliferation via Nr4a1 and Nr4a3 nuclear receptors. Proc Natl Acad Sci U S A. 2014;111(14):5242–5247. doi:10.1073/pnas.1320953111
- Robertson RP. Chronic oxidative stress as a central mechanism for glucose toxicity in pancreatic islet beta cells in diabetes. J Biol Chem. 2004;279(41):42351–42354. doi:10.1074/jbc.R400019200
- Robertson RP, Harmon JS. Diabetes, glucose toxicity, and oxidative stress: a case of double jeopardy for the pancreatic islet β cell. Free Radic Biol Med. 2006;41(2):177–184. doi:10.1016/j.freeradbiomed.2005.04.030
- Mathis D, Vence L, Benoist C. β-Cell death during progression to diabetes. Nature. 2001;414(6865):792–798. doi:10.1038/414792a
- Marinho HS, Real C, Cyrne L, et al. Hydrogen peroxide sensing, signaling and regulation of transcription factors. Redox Biol. 2014;2:535–562. doi:10.1016/j.redox.2014.02.006
- Alam J, Cook JL. How many transcription factors does it take to turn on the heme oxygenase-1 gene? Am J Respir Cell Mol Biol. 2007;36(2):166–174. doi:10.1165/rcmb.2006-0340TR
- Shimizu Y, Miyakura R, Otsuka Y. Nuclear receptor subfamily 4, group A, member 1 inhibits extrinsic apoptosis and reduces caspase-8 activity in H2O2-induced human HUC-F2 fibroblasts. Redox Rep. 2015;20(2):81–88. doi:10.1179/1351000214Y.0000000109
- Safe S, Jin UH, Morpurgo B, et al. Nuclear receptor 4A (NR4A) family - orphans no more. J Steroid Biochem Mol Biol. 2016;157:48–60. doi:10.1016/j.jsbmb.2015.04.016
- Lee SO, Jin UH, Kang JH, et al. The orphan nuclear receptor NR4A1 (Nur77) regulates oxidative and endoplasmic reticulum stress in pancreatic cancer cells. Mol Cancer Res. 2014;12(4):527–538. doi:10.1158/1541-7786.MCR-13-0567
- Volakakis N, Kadkhodaei B, Joodmardi E, et al. NR4A orphan nuclear receptors as mediators of CREB-dependent neuroprotection. Proc Natl Acad Sci U S A. 2010;107(27):12317–12322. doi:10.1073/pnas.1007088107
- McCluskey JT, Hamid M, Guo-Parke H, et al. Development and functional characterization of insulin-releasing human pancreatic beta cell lines produced by electrofusion. J Biol Chem. 2011;286(25):21982–21992. doi:10.1074/jbc.M111.226795
- Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65(1-2):55–63. doi:10.1016/0022-1759(83)90303-4
- Oba R, Kudo Y, Sato N, et al. A new method of competitive reverse transcription polymerase chain reaction with SYBR Gold staining for quantitative analysis of mRNA. Electrophoresis. 2006;27(14):2865–2868. doi:10.1002/elps.200500568
- Chaffey JR, Young J, Leslie KA, et al. Investigation of the utility of the 1.1B4 cell as a model human beta cell line for study of persistent enteroviral infection Sci Rep. 2021;11(1):15624.
- St-Pierre J, Drori S, Uldry M, et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127(2):397–408. doi:10.1016/j.cell.2006.09.024
- Maxwell MA, Muscat GE. The NR4A subgroup: immediate early response genes with pleiotropic physiological roles. Nucl Recept Signal. 2006;4:nrs.04002. doi:10.1621/nrs.04002
- Robertson RP. Antioxidant drugs for treating beta-cell oxidative stress in type 2 diabetes: glucose-centric versus insulin-centric therapy. Discov Med. 2010;9(45):132–137.
- Nomi Y, Shimizu S, Sone Y, et al. Isolation and antioxidant activity of zeylaniin A, a new macrocyclic ellagitannin from Syzygium zeylanicum leaves. J Agric Food Chem. 2012;60(41):10263–10269. doi:10.1021/jf302977n