ABSTRACT
Among hospitalized patients worldwide, infections caused by multidrug-resistant (MDR) bacteria are a major cause of morbidity and mortality. This study aimed to isolate MDR bacteria from five intensive care units (ICUs) at Tripoli University Hospital (TUH). A prospective cross-sectional study was conducted over a seven-month period (September 2022 to March 2023) across five ICUs at TUH. A total of 197 swabs were collected from Patients’, healthcare workers’ and ICUs equipment. Samples collected from patients were nasal swabs, oral cavity swabs, hand swabs, sputum specimens, skin swabs, umbilical venous catheter swabs, and around cannula. Swabs collected from health care workers were nasal swabs, whereas ICUs equipment’s samples were from endotracheal tubes, oxygen masks, and neonatal incubators. Identification and antimicrobial susceptibility test was confirmed by using MicroScan auto SCAN 4 (Beckman Coulter). The most frequent strains were Gram negative bacilli 113 (57.4%) with the predominance of Acinetobacter baumannii 50/113 (44%) followed by Klebsiella pneumoniae 44/113 (40%) and Pseudomonas aeruginosa 6/113 (5.3%). The total Gram positive bacterial strains isolated were 84 (42.6%), coagulase negative Staphylococci 55 (66%) with MDRs (89%) were the most common isolates followed by Staphylococcus aureus 15 (17.8%). Different antibiotics were used against these isolates; Gram- negative isolates showed high resistance rates to ceftazidime, gentamicin, amikacin and ertapenem. A. baumannii were the most frequent MDROs (94%), and the highest resistance rates in Gram-positive strains were observed toward ampicillin, oxacillin, ampicillin/sulbactam and Cefoxitin, representing 90% of total MDR Gram-positive isolates. ESBL and MRS were identified in most of strains. The prevalence of antibiotic resistance was high for both Gram negative and Gram positive isolates. This prevalence requires strict infection prevention and control intervention, continuous monitoring, implementation of effective antibiotic stewardship, immediate, concerted and collaborative action to monitor its prevalence and spread in the hospital.
1. Introduction
Antimicrobial resistance (AMR) is a global health challenge facing health care professionals worldwide, it is responsible for morbidity and mortality and burdens healthcare systems [Citation1], Health care associated Infections are the leading cause of surgical failure, transplant rejection, with antibiotic-resistant organisms responsible for increased morbidity and mortality [Citation1]. Although life-threatening bacterial infections have been treated with antibiotics for decades and greatly improved the well-being and survival of human beings, bacteria have taken a step forward by resisting antibiotics through various mechanisms [Citation2,Citation3]. Furthermore, report on antimicrobial resistance threats released by center for disease prevention and control (CDC) in 2019 shows that annually; 2.8 million cases of infections caused by antibiotic-resistant bacteria is recorded with 35.000 deaths in the US [Citation4]. The rising phenomenon of antibiotic resistance is a direct result of irrational antibiotic use in humans, animals, and agriculture. This is particularly evident in developing countries, where antibiotics are easily and un-regulatory accessible. The resistance is acquired either naturally or acquired by transfer of resistance genes. The continual use of different drugs has exerted selection pressure on bacteria, transforming them into MDR microorganisms [Citation5]. The explosion of MDR bacteria has far-reaching consequences for both clinical outcomes and economic burdens. Nevertheless, the thoughtless use of antibiotics during the COVID-19 pandemics further hastened bacterial resistance, leading to a dramatic increase in morbidity and mortality worldwide [Citation6]
World Health Organization (WHO) published its list of antibiotic-resistant ‘priority pathogens’ that pose the greatest threat to human health [Citation7]. The list is divided into three categories according to the urgency of needs for new antibiotics: critical, high and medium priorities. The most critical group includes Acinetobacter, Pseudomonas and Enterobactereales, the former include carbapenems resistant and Extended Spectrum Beta-lactamase (ESBL) producing Enterobacteriaceae. These microorganisms cause serious and often fatal infections particularly in healthcare settings. The high and medium group contain other Gram positive and negative bacteria such as vancomycin-resistant Enterococcus faecium, methicillin-resistant S. aureus (MRSA), vancomycin- resistant S. aureus (VRSA) and Neisseria gonorrhoeae, cephalosporins-resistant Enterobactereales [Citation7]. MRSA and vancomycin-resistant Enterococci (VRE) are the most common MDR Gram-positive bacteria in hospitals. MRSA infections appear to be declining, and therapeutic options are available for both MRSA and VRE infections. On the other hand, MDR Gram-negative bacteria, including Enterobacteriaceae, Pseudomonas, and Acinetobacter, continue to pose a significant threat [Citation8].
By identifying antibiotic-resistant bacteria colonizing hospitalized patients and healthcare workers, or contaminating equipment, we can implement appropriate contact precautions to prevent the spread of these infections during patient hospitalization. Healthcare facilities are a breeding ground for antibiotic-resistant bacteria, which can easily transfer from the environment or healthcare personnel to patients [Citation9]. Identifying these bacteria and their transmission routes (direct contact with colonized/infected patients or via healthcare worker hands) is crucial. This knowledge allows us to implement appropriate contact precautions and prevent infections during patient hospitalization.
In addition, early identification of antibiotic-resistant pathogens and appropriate targeted antibiotic therapy are the most important factors in reducing mortality from antibiotic resistant pathogens [Citation4]. Growing and widespread emergence of antibiotic resistant bacteria has forced researchers to find alternative treatments for antibiotic resistant bacteria. However, despite global efforts, there are significant gaps in epidemiological data, particularly in low- and middle-income countries (LMICs) including Libya.
There is a paucity data on antibiotic resistant bacteria and antibiotic use in Libya due to the lack of surveillance system for antimicrobial resistance by the Ministry of Health and National Centre for Disease Prevention and Control in Libya along with the low number of studies compared to other countries.
Studies in Libya found carbapenem-resistant bacteria in hospitals. Tripoli metallo- β -lactamase (TMB-1) was detected in Achromobacter xylosoxidans isolated from the hospital environment of Tripoli Central Hospital and was found in a class 1 integron of 3kb [Citation10]. This included Pseudomonas aeruginosa and Acinetobacter baumannii, which carried genes like VIM-2, OXA-23, and NDM-1 [Citation11,Citation12]. These bacteria were present in various hospital areas, including intensive care and burn units. Additionally, carbapenem-resistant Klebsiella pneumoniae carrying OXA-48 was detected [Citation13]. Worryingly, carbapenem and colistin-resistant bacteria were also isolated from COVID-19 patients in Libya [Citation14]. ESBL and MBL producing Gram-negative bacteria were isolated from operating theaters and ICU of Sabha Medical Center, Sabha city, Libya [Citation15,Citation16].
The current study was conducted prospectively focusing on the prevalence of MDR bacteria among hospitalized patients, healthcare workers and equipment used on patients. The main goal of our study was to determine the most prevalent MDR bacteria and their antibiotic susceptibility pattern trying to fill the gap of knowledge regarding the most frequently encountered MDR microorganisms in health care settings. Our study will highlight the serious concern of antibiotic resistant bacteria and the extent of the problem within the ICUs of the studied health care facility, and will provide useful information on the susceptibility patterns and may guide clinicians in choosing appropriate treatment through constructing antibiogram for resistant bacteria depending on the result of antibiotic susceptibility. The results of the study will be used to inform the Libyan health care system regarding the extent of AMR in a Libyan health care setting.
2. Materials and methods
2.1. Study design and patients
This study was conducted in the microbiology department at the Libyan Biotechnology Research Center in Tripoli, Libya from September 2022 to March 2023. Clinical samples were collected from medical staff, equipment, and patients hospitalized in Tripoli University Hospital (TUH) ‘s five intensive care units (ICUs). This hospital is the largest hospital in Libya with 1200 beds. The ICUs were; medical ICU (MICU) with 15-bed capacity, neonatal ICU (NICU) with 16-bed capacity, cardiology pediatric care unit (CPCU) with 12-bed capacity, surgical ICU (SICU) with 8-bed capacity, and pediatric ICU (PICU) with 9-bed capacity. Patient’s samples were taken after their informed oral consent.
2.2. Samples collection
This is a cross-sectional study. A total of 197 swabs were collected once from patients, medical staff and ICUs equipment’s using moistened (0.9% w/v physiological saline) sterile cotton swabs. Multiple swabs from different sites were sometimes taken from the same patient according to their clinical presentation. Samples collected from patients were; nasal swabs, oral cavity swabs, hand swabs, sputum specimens, swabs from ulcer, surgical wounds, umbilical venous catheter swabs, and around cannula. Swabs collected from health care workers were nasal swabs whereas ICUs disposables and equipment’s samples were from; endotracheal tubes, oxygen masks, and neonatal incubators.
2.3. Isolation of bacterial strains
Bacterial swabs were inoculated in Brain Heart Infusion (BHI) and incubated at 37°C for 18–24 h. The samples were then inoculated on MacConkey and MSA agar (Oxoid, UK) and incubated at 37°C for 18–24 h. The initial identification of the bacterial isolates was performed using morphological analysis, Gram staining, and microscopy in accordance with microbiological standards. Each isolate was stored in Trypticase soy broth with 20% glycerol at a temperature of −70°C.
2.4. Identification and antimicrobial susceptibility testing
The identification and antimicrobial susceptibility of the bacterial isolates were determined using a micro scan auto-SCAN 4 panel (Beckman Coulter). The tested antibiotics for Gram-negative bacteria were; amikacin, ampicillin, amoxicillin/clavulanate, ampicillin/sulbactam, piperacillin/tazobactam, cefoxitin, cefazolin, cefotaxime, cefotaxime/clavulanate, ceftazidime, ceftazidime/clavulanate, cefuroxime, cefepime, aztreonam, imipenem, meropenem, ertapenem, gentamicin, tobramycin, ciprofloxacin, levofloxacin, moxifloxacin, norfloxacin, nitrofurantoin, colistin, tigecycline, and trimethoprim/sulphamethoxazole. Extended spectrum beta lactamase (ESBL) production was determined as well as using a Microscan auto-SCAN 4 panel (Beckman Coulter).
The tested antibiotics panel for Gram-positive bacteria were; penicillin, ampicillin, ampicillin/sulbactam, amoxicillin/clavulanate, oxacillin, cefoxitin screen, ceftriaxone, ciprofloxacin, levofloxacin, gentamicin, clindamycin, erythromycin, tetracycline, vancomycin, daptomycin, linezolid, nitrofurantoin, rifampin, quinupristin/dalfopristin teicoplanin, and trimethoprim/sulfamethoxazole.
3. Results
The results showed that the 197 swabs yielded 197 isolates, they were from patients’ (131), healthcare workers’ (44) and medical equipment’s (22), (, ). The distribution of bacterial isolates among the five ICUs were as follows; (NICU = 35), (MICU = 66), (PICU = 17), (SICU = 40), and (CPCU = 39) as shown in .
Figure 1. Prevalence of pathogenic bacteria isolated from patients, healthcare workers and ICUs medical equipment’s in TUH ICUs. NICU (neonatal ICU), MICU (medical ICU), PICU (pediatric ICU), SICU (surgical ICU) and CPCU (cardiology pediatric care unit).
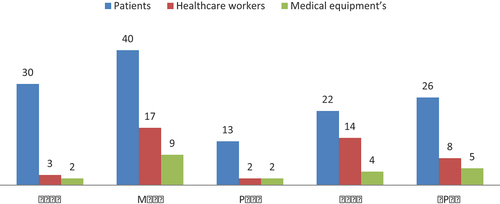
Table 1. Distribution of bacterial isolates among Tripoli University Hospital ICUs.
The prevalence of Gram-negative and Gram-positive isolates among the ICUs is illustrated in . The results showed that Gram-negative isolates were the most frequent strains (113; 57.4%). The predominant strains were A. baumannii 50/113 (44%), followed by K. pneumoniae 46/113 (40%), P. aeruginosa 6/113 (5.3%), Chryseobacterium indologenes 3/113 (2.6%), Serratia fonticola 3/113 (2.6%), Myroides species 2/113 (1.8%), and one strain of Enterobacter aerogenes, Pantoea agglomerans, and Pasteurella aerogenes.
Table 2. Prevalence of Gram-negative and Gram-positive isolates in Tripoli University Hospital ICUs.
The prevalence of Gram-positive isolates among the ICUs was 84/197 (42.6%), with the commonest strains being S. epidermidis 42/84 (50%), followed by S. aureus 15/84 (17.8%), S. haemolyticus 9/84 (10.7%), S. sciuri 4/84 (4.8%), S. hominis subsp. homoinis 2/84 (2.3%) and one strain of S. xylosus, S. auricularis and S. simulans. In general, Coagulase Negative Staphylococci (CONS) strains were the most common isolates, 69/84 (82.2%), followed by Coagulase Positive Staphylococci strains 15/84 (17.8%).
Antimicrobial sensitivity was done using a Micro Scan auto SCAN 4 (Beckman Coulter). As shown in , most Gram negative isolates showed the highest resistance to ceftazidime (96.5%), gentamicin (95%), amikacin (92%), ertapenem (91.2%), ampicillin/sulbactam (90%), imipenem (89.5%) and Pipracillin/Tazobactam (89%). In contrast, the least resistance rate was documented for cefazolin (23%), cefoxitin (26.5%) and moxifloxacin (32%). Extended spectrum β-lactamase (ESBL) was detected by micro scan auto-SCAN 4 panel (Beckman Coulter) using aztreonam, cefotaxime and ceftazidime. However, A. baumannii were the most frequent MDROs (94%), followed by K. pneumoniae (90%) and P. aeruginosa (60%) of the whole MDR Gram- negative isolates in the study. Moreover, ESBL was identified in most of Gram negative isolates 97/113 (85.8%) ().
Table 3. Antibiotic resistance pattern of Gram-negative strains in TUH ICUs.
Among Gram positive strains, the highest resistance rates were toward ampicillin (100%), oxacillin (95%), ampicillin/sulbactam (94%), amoxicillin/clavulanate (93%) and ceftriaxone (93%), while the least resistance rate was toward vancomycin (69%), quinupristin/dalfopristin (69%) and rifampin (75%). Most of Gram positive strains were MDR (84%) and MRS 74/84 (88%). However, S. epidermidis were the most frequent MDROs (96%) followed by S. aureus (92%). Additionally, MRSA and MRSE were observed in most strains 18% and 50%, respectively. Notably, resistance to colistin was observed in K. pneumoniae, A.baumannii and P. aeruginosa. In general, Gram negative and Gram positive strains that were isolated from PICU, CPICU and SICU were more resistant to majority of antibiotics compared to other ICUs ().
Table 4. Antibiotic resistance pattern of Gram-positive strains in TUH ICUs.
4. Discussion
In intensive care units, hospitalized patients are more susceptible to nosocomial pathogens, in particular multidrug-resistant bacteria. These bacteria can spread from patients’ health care workers and ICU medical equipment and vice versa, leading to hospital outbreaks by cross-infection and/or cross contamination [Citation17]. Therefore, the main objective of this study was to determine the MDR profile of pathogenic bacteria isolated from specimens collected and their antibiotic susceptibility patterns in various intensive care units at the TUH.
The incidence of infections caused by multi-drug-resistant pathogens is rising around the world. Several studies presented different frequencies of MDR among microorganisms in many healthcare settings. In the present study, A. baumannii strains were the most frequent MDROs representing (94%) followed by K. pneumonia strains (90%) of the whole MDR gram negative patient [Citation18] isolates. Our clinical samples results are consistent with several studies. For instance, in Egypt a rate of 57% MDR bacteria were reported in critically ill elderly patients, with Gram-negative bacteria comprised the majority, representing 72% of total MDR isolates [Citation19]. In another study, the dominance of MDR Gram-negative over Gram-positive bacteria has been shown from infected wounds [Citation20]. Furthermore, in a study by Abdelkader MM et. al. on cerebrospinal fluid; 36.6% of the isolates were MDR of which 69.2% were Gram-negatives and 30.8% were Gram-positives [Citation20]. Additionally, in Sudan, 80% of K. pneumoniae were reported as MDR [Citation21], and one more study found that there was a high prevalence of MDR Enterobacteriaceae; mainly ESBL-positive E. coli and K. pneumoniae [Citation22], Whereas 18% of P. aeruginosa isolates were MDR in a recent work published in 2023 [Citation23], and 89.2% of S. aureus were MDR, of them 67.6% were methicillin resistant [Citation24,Citation25].
In another study in Tunisia, a MDR rate of 44% was detected represented mainly by A. baumannii, ESBL producing Enterobacteriaceae and P. æruginosa [Citation26]. An Investigation of the carriage of MDR bacteria among hospitalised paediatric patients found that, there was a 7.84% acquisition rate of MDR bacteria on discharge [Citation27]. In Libya, due to lack of national surveillance system and few published studies, little data are available on antimicrobial resistance. Seventy-eight percent (78%) of the GNB exhibit various MDR phenotypes, of these, K. pneumoniae showed the highest rates of MDRs (91%) in a study published in 2017 [Citation28]. Rate of ESBLs among E. coli ranged from 5%-20% and from 11%-18% for K. pneumoniae in some studies from Libya [Citation29,Citation30]. These results are nearly in agreement with the our results in which the most common isolates were Gram-negative strains 113 (57.4%) with the predominance of A. baumannii 50/113 (44%) followed by K. pneumoniae 44/113 (40%), and P. aeruginosa 6/113 (5.3%). Gram positive strains were 84 (42.6%) isolates, CoNS strains represented 82.2% as the most commonly isolated strains followed by S. aureus (17.8%).
The current findings are also consistent with the recent study that demonstrated the emergence of MDR-gram-negative bacteria in Tripoli and Benghazi hospitals from clinical and non-clinical specimens. K. pneumoniae was found to be the most frequently isolated strain being disseminated in hospitals, followed by E. coli and A. baumannii [Citation31]. Moreover, methicillin resistance (MR) was detected in 35.3% of S. aureus and in 75% of coagulase-negative Staphylococci (CNS). However, in our study, coagulase-negative Staphylococci isolates (82.2%) were the most common strains, followed by S. aureus (17.8%), as shown in and . In general, Acinetobacter strains were the most common pathogens isolates in this investigation, which was consistent with a previous study [Citation32,Citation33]. Conversely, it has been demonstrated in recent study [Citation34,Citation35] that Klebsiella spp. are important causative agent of nosocomial infections in intensive care units. This discrepancy could be explained by variations in the patient’s immune system, immunological status, nutritional state, and geographical location. In a worldwide research of the incidence of infections in intensive care units, ‘Infections in Intensive Care Study,’ Acinetobacter was similarly found to be the most common pathogen recovered from patients [Citation32,Citation36].
Some Gram-negative species are naturally resistant to colistin. Examples include Neisseria meningitidis, Burkholderia species, and Proteus mirabilis [Citation37,Citation38]. In this study, Klebsiella pneumoniae, Acinetobacter baumannii, and Pseudomonas aeruginosa have acquired resistance to this antibiotic. Colistin resistance is a significant concern because colistin-resistant bacteria emerged due to uncontrolled use of this last line antibiotic for multidrug-resistant Gram-negative bacteria [Citation39,Citation40]. The current findings found that the prevalence of antibiotic resistance in five ICUs at TUH was high for both Gram negative and Gram positive isolates. However, using a program consisting of persuasive stewardship measures, could be useful to control drug-resistant bacteria in a hospital setting [Citation41]. Furthermore, the rises registered recommend that efforts to control the spread of pathogenic and antibiotic-resistant bacteria could require to be strengthened, such as through improved infection control practices and proper use of antibiotics. Moreover, it’s essential to continue monitoring these trends to monitor the success of therapies and spot new trends in resistance [Citation42]. Therefore, these persuasive stewardship measures can be used to reduce MDR and nosocomial infections within our hospital setting.
This study has several limitations. All analyzed strains are not necessarily associated to a clinical relevant infection. However, it may reflect a clinical relevant problem of antimicrobial resistance in the 5 ICUs of Tripoli University Hospital. Also, analyzing bacteria from a single sampling point limits understanding of how these populations change over time. Additionally, not differentiating between staff and patient sources limits identification of transmission routes. Additionally, the study did not analyze particular resistance genes. Despite these limitations, our findings highlight the potential risk posed by the presence of these bacteria and emphasize the importance of ongoing surveillance and infection control measures within the ICUs of Tripoli University Hospital.
5. Conclusions
This study demonstrated the alarming occurrence of multi-drug-resistant bacteria in various intensive care units at TUH, which is considered the largest hospital in Libya with 1200 beds. In this study, Gram-negative isolates were the most frequent strains; A. baumannii complex/haemolyticus followed by K. pneumoniae; and Coagulase Negative Staphylococci (CONS) were the major pathogens prevalent strains in the five ICUs. This prevalence and high resistance rate requires immediate, concerted and collaborative action to monitor its prevalence and spread throughout the Libyan hospitals.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Rana MM, Grossi P, Huprikar S. Multidrug-resistant organisms in Solid Organ Transplantation. In: Manuel O Ison M, editors. Infectious diseases in Solid-Organ transplant recipients: a practical approach [internet]. Cham: Springer International Publishing; 2019 [cited 2024 Apr 8]. p. 145–8. doi: 10.1007/978-3-030-15394-6_11
- Hutchings MI, Truman AW, Wilkinson B. Antibiotics: past, present and future. Curr Opin Microbiol. 2019;51:72–80. doi: 10.1016/j.mib.2019.10.008
- Khan AA, Manzoor KN, Sultan A, et al. Pulling the Brakes on Fast and furious multiple drug-resistant (MDR) bacteria. Int J Mol Sci [Internet]. 2021 cited 2024 Apr 8;22(2):859. doi: 10.3390/ijms22020859
- CDC. The biggest antibiotic-resistant threats in the U.S. [internet]. Centers for Disease Control and Prevention. 2022 [cited 2024 Apr 8]. Available from: https://www.cdc.gov/drugresistance/biggest-threats.html
- Uchil RR, Kohli GS, Katekhaye VM, et al. Strategies to combat antimicrobial resistance. J Clin Diagn Res [Internet]. 2014 [cited 2024 Apr 8];8:ME01–ME04. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4149102/
- Rizvi SG, Ahammad SZ. COVID-19 and antimicrobial resistance: a cross-study. Sci Total Environ [Internet]. 2022 [cited 2024 Apr 8];807:150873. Available from: https://www.sciencedirect.com/science/article/pii/S0048969721059519
- Tacconelli E, Carrara E, Savoldi A, et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18(3):318–327. doi: 10.1016/S1473-3099(17)30753-3
- Al-Said H, Alghamdi A, Ashgar S, et al. Isolation and detection of drug-resistant bacterial pathogens in postoperative wound infections at a tertiary care hospital in Saudi Arabia. Saudi j med med sci [Internet]. 2023 cited 2024 Apr 9;11(3):229. Available from: https://pubmed.ncbi.nlm.nih.gov/37533663/
- Russotto V, Cortegiani A, Fasciana T, et al. What healthcare workers should know about environmental bacterial contamination in the intensive care unit. Biomed Res Int. 2017;2017:1–7. doi: 10.1155/2017/6905450
- El Salabi A, Borra PS, Toleman MA, et al. Genetic and biochemical characterization of a novel metallo-β-lactamase, TMB-1, from an Achromobacter xylosoxidans strain isolated in Tripoli, Libya. Antimicrob Agents Chemother [Internet]. 2012 [cited 2024 Apr 9];56(5):2241–2245. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3346670/
- Mathlouthi N, Areig Z, Al Bayssari C, et al. Emergence of carbapenem-resistant Pseudomonas aeruginosa and acinetobacter baumannii clinical isolates collected from some Libyan hospitals. Microb Drug Resist. 2015;21(3):335–341. doi: 10.1089/mdr.2014.0235
- Mathlouthi N, El Salabi AA, Ben Jomàa-Jemili M, et al. Early detection of metallo-β-lactamase NDM-1- and OXA-23 carbapenemase-producing acinetobacter baumannii in Libyan hospitals. Int J Antimicrob Agents. 2016;48(1):46–50. doi: 10.1016/j.ijantimicag.2016.03.007
- Mathlouthi N, Al-Bayssari C, El Salabi A, et al. Carbapenemases and extended-spectrum β-lactamases producing enterobacteriaceae isolated from Tunisian and Libyan hospitals. J Infect Dev Ctries. 2016;10(7):718–727. doi: 10.3855/jidc.7426
- Slimene K, Ali AA, Mohamed EA, et al. Isolation of carbapenem and colistin resistant gram-negative bacteria colonizing immunocompromised SARS-CoV-2 patients admitted to some Libyan hospitals. Microbiol Spectr [Internet]. 2023 [cited 2024 Apr 9];11(3):e02972–22. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10269485/
- Alrbee AA, Elzen A, Salabi AAE. Isolation of multi-drug resistant gram-negative bacteria producing extended spectrum beta lactamases in operating theaters of sebha medical center, Sebha, Libya. JOPAS [Internet]. 2019 [cited 2024 Apr 9];18. Available from: https://sebhau.edu.ly/journal/jopas/article/view/313
- Alrbee AA, Elzen A, Salabi AAE. Detection of multi-drugs resistant gram-negative bacilli bacteria producing metallo beta lactamases in intensive care unit of sebha medical center, Sebha, Libya. JOPAS [Internet]. 2022 [cited 2024 Apr 9];21:261–269. Available from: https://sebhau.edu.ly/journal/jopas/article/view/2205
- Galvin S, Dolan A, Cahill O, et al. Microbial monitoring of the hospital environment: why and how? J Hosp Infect. 2012;82(3):143–151. doi: 10.1016/j.jhin.2012.06.015
- Elsorady KE, Hassan RA, Abdelhamid DH, et al. Antibiotics use and its association with multi-drug resistance in a tertiary care geriatrics hospital in Egypt. J Infect Dev Ctries. 2022;16(12):1860–1869. doi: 10.3855/jidc.17257
- Hassan MA, Abd El-Aziz S, Elbadry HM, et al. Prevalence, antimicrobial resistance profile, and characterization of multi-drug resistant bacteria from various infected wounds in North Egypt. Saudi J Biol Sci. 2022;29(4):2978–2988. doi: 10.1016/j.sjbs.2022.01.015
- Abdelkader MM, Aboshanab KM, El-Ashry MA, et al. Prevalence of MDR pathogens of bacterial meningitis in Egypt and new synergistic antibiotic combinations. PLOS ONE. 2017;12(2):e0171349. doi: 10.1371/journal.pone.0171349
- Osman EA, Yokoyama M, Altayb HN, et al. Klebsiella pneumonia in Sudan: multidrug resistance, polyclonal dissemination, and virulence. Antibiotics. 2023;12(2):233. doi: 10.3390/antibiotics12020233
- Moglad EH. Antibiotics profile, prevalence of extended-spectrum beta-lactamase (ESBL), and multidrug-resistant enterobacteriaceae from different clinical samples in Khartoum State, Sudan. Int J Microbiol. 2020;2020:8898430. doi: 10.1155/2020/8898430
- Hussain MA, Mohamed MS, Altayb HN, et al. Comparative genomic analysis of multi-drug resistant Pseudomonas aeruginosa sequence type 235 isolated from Sudan. Microorganisms. 2023;11(6):1432. doi: 10.3390/microorganisms11061432
- Siddig LA, Bayoumi M, Elhadi N. Sociodemographic distributions and molecular characterization of colonized Enterococcus faecium isolates from locality hospitals in Khartoum, Sudan. PeerJ. 2023;11:e16169. doi: 10.7717/peerj.16169
- Moglad EH, Altayb HN. Antibiogram, prevalence of methicillin-resistant and multi-drug resistant staphylococcus spp. In different clinical samples. Saudi J Biol Sci. 2022;29(12):103432. doi: 10.1016/j.sjbs.2022.103432
- Frigui S, Bourbiaa Y, Mokline A, et al. Nosocomial bacteremia in burns: clinical and bacterial epidemiology. Ann Burns Fire Disasters. 2021;34(1):10–17.
- Tfifha M, Ferjani A, Mallouli M, et al. Carriage of multidrug-resistant bacteria among pediatric patients before and during their hospitalization in a tertiary pediatric unit in Tunisia. Libyan J Med. 2018;13(1):1419047. doi: 10.1080/19932820.2017.1419047
- Elramalli A, Almshawt N, Ahmed MO. Current problematic and emergence of carbapenemase-producing bacteria: a brief report from a Libyan hospital. Pan Afr Med J. 2017;26:180. doi: 10.11604/pamj.2017.26.180.9637
- Zorgani A, Almagatef A, Sufya N, et al. Detection of CTX-M-15 among uropathogenic Escherichia coli isolated from five Major hospitals in Tripoli, Libya. Oman Med J. 2017;32(4):322–327. doi: 10.5001/omj.2017.61
- Abujnah AA, Zorgani A, Sabri MAM, et al. Multidrug resistance and extended-spectrum β-lactamases genes among Escherichia coli from patients with urinary tract infections in Northwestern Libya. Libyan J Med. 2015;10(1):26412. doi: 10.3402/ljm.v10.26412
- Salabi AAE. Characterisation of antibiotic resistance mechanisms in Gram-negative bacteria from Tripoli and Benghazi, Libya [internet]. Published by ProQuest LLC 2013; 2011 [cited 2024 Apr 9]. Available from: https://orca.cardiff.ac.uk/id/eprint/55122/1/U585511.pdf
- Bhandari P, Thapa G, Pokhrel BM, et al. Nosocomial isolates and their drug resistant pattern in ICU patients at national institute of neurological and allied sciences, Nepal. Int. J. Microbiol. [Internet] 2015 [cited 2024 Apr 9];2015:1–6. Available from: http://www.hindawi.com/journals/ijmicro/2015/572163/
- Patwardhan RB, Dhakephalkar PK, Niphadkar KB, et al. A study on nosocomial pathogens in ICU with special reference to multiresistant acinetobacter baumannii harbouring multiple plasmids. Indian J Med Res. 2008;128:178–187.
- Radji M, Fauziah S, Aribinuko N. Antibiotic sensitivity pattern of bacterial pathogens in the intensive care unit of Fatmawati Hospital, Indonesia. Asian Pac J Trop Biomed. 2011;1(1):39–42. doi: 10.1016/S2221-1691(11)60065-8
- Mythri H, Kashinath K. Nosocomial infections in patients admitted in intensive care unit of a tertiary health center, India. Ann Med Health Sci Res. 2014;4(5):738–741. doi: 10.4103/2141-9248.141540
- Vincent J-L, Rello J, Marshall J, et al. International study of the prevalence and outcomes of infection in intensive care units. JAMA. 2009;302(21):2323–2329. doi: 10.1001/jama.2009.1754
- Torres DA, Seth-Smith HMB, Joosse N, et al. Colistin resistance in Gram-negative bacteria analysed by five phenotypic assays and inference of the underlying genomic mechanisms. BMC Microbiology [Internet]. 2021 [cited 2024 Apr 8];21(1):321. Available from. doi: 10.1186/s12866-021-02388-8
- Olaitan AO, Morand S, Rolain J-M. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol [Internet]. 2014 [cited 2024 Apr 9];5. Available from: https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2014.00643/full
- Paterson DL, Harris PNA. Colistin resistance: a major breach in our last line of defence. Lancet Infect Dis [Internet]. 2016 [cited 2024 Apr 12];16(2):132–133. Available from: https://www.sciencedirect.com/science/article/pii/S1473309915004636
- Chew KL, La M-V, Lin RTP, et al. Colistin and polymyxin B susceptibility testing for carbapenem-resistant and mcr-Positive Enterobacteriaceae: comparison of sensititre, MicroScan, Vitek 2, and etest with broth microdilution. J Clin Microbiol. 2017;55(9):2609–2616. doi: 10.1128/JCM.00268-17
- Scaglione V, Reale M, Davoli C, et al. Prevalence of antibiotic resistance over time in a third-level university hospital. Microbial Drug Resist [Internet]. 2022 [cited 2024 Apr 9];28(4):425–435. Available from: https://www.liebertpub.com/doi/10.1089/mdr.2021.0109
- Cerini P, Meduri FR, Tomassetti F, et al. Trends in antibiotic resistance of nosocomial and community-acquired infections in Italy. Antibiotics. 2023;12(4):651. doi: 10.3390/antibiotics12040651

