ABSTRACT
We argue that a paradigm shift is needed in the analysis of phage DNA packaging. We then test a prediction of the following paradigm shift-engendering hypothesis. The motor of phage DNA packaging has two cycles: (1) the well-known packaging ATPase-driven (type 1) cycle and (2) a proposed back-up, shell expansion/contraction-driven (type 2) cycle that reverses type 1 cycle stalls by expelling accidentally packaged non-DNA molecules. We test the prediction that increasing the cellular concentration of all macromolecules will cause packaging-active capsids to divert to states of hyper-expansion and contraction. We use a directed evolution-derived, 3-site phage T3 mutant, adapted to propagation in concentrated bacterial cytoplasm. We find this prediction correct while discovering novel T3 capsids previously obscure.
The motors of phage DNA packaging are useful to model eukaryotic motors in part because in vivo-generated phage motor states can be analyzed. Eukaryotic motors (e.g., actin-myosin motors) are analyzed only after assembly in vitro (reviewsCitation1-3).
However, analysis of phage DNA packaging ( for the related phages, T3 and T7) appears to need a paradigm shift. Hypotheses typically don't fit all data.Citation4 A packaging ATPase-derived power stroke is demonstrated in vitro, both in practiceCitation5 and in theory.Citation4 But, the power stroke of phage phi29 varies in length, which raises the unanswered question of whether the capsid's shell participates in packaging.Citation5 This question also arises from effects on neighboring shell structure of phi29 packaging motor components.Citation6
Figure 1. DNA packaging of the related phages, T3 and T7. (A) DNA packaging in vivo is illustrated with capsid II participating in the type 1 and proposed type 2 cycles (reviewCitation4). (B) U-NLD capsid II and particles from neighboring fractions [fraction density (g/ml) indicated at the top] are analyzed by SDSPAGE in a 9% polyacrylamide separating gel. The lane labeled WT-CII has wild type T3 NLD capsid II; the lane labeled U-NLD has T3Su-1 U-NLD capsid II. The arrow indicates the direction of electrophoresis; arrowheads indicate origins of electrophoresis. Some gp9 scaffolding protein is present in capsid II. The gp9/gp10 ratio is, however, higher for capsid I. For reasons not known, wild type gp8 formed a doublet band, which it usually does not. (C) EM of U-NLD capsid II with the procedure of references Citation27 and Citation28.
![Figure 1. DNA packaging of the related phages, T3 and T7. (A) DNA packaging in vivo is illustrated with capsid II participating in the type 1 and proposed type 2 cycles (reviewCitation4). (B) U-NLD capsid II and particles from neighboring fractions [fraction density (g/ml) indicated at the top] are analyzed by SDSPAGE in a 9% polyacrylamide separating gel. The lane labeled WT-CII has wild type T3 NLD capsid II; the lane labeled U-NLD has T3Su-1 U-NLD capsid II. The arrow indicates the direction of electrophoresis; arrowheads indicate origins of electrophoresis. Some gp9 scaffolding protein is present in capsid II. The gp9/gp10 ratio is, however, higher for capsid I. For reasons not known, wild type gp8 formed a doublet band, which it usually does not. (C) EM of U-NLD capsid II with the procedure of references Citation27 and Citation28.](/cms/asset/63bc9d4e-0145-46f2-964a-6194b566eda8/kbac_a_1268664_f0001_oc.gif)
In addition, (1) single-molecule studies of phage T4 show that one inactive subunit in a packaging ATPase ring (green in ) does not inactivate packaging (referenceCitation7 and personal communication from V. Rao), in contrast to what most theories require,Citation4 (2) predictions of previous theories are generally incorrect (reviews.Citation4,8-10), (3) no phage is known to package host cytoplasmic components (reviewsCitation4,8-10), even though packaging-associated exit of scaffolding proteins indicates hole formation for several phages, including the related phages, T3 and T7 (), and (4) old paradigms do not explain the observation, for phages phi29Citation11, T3Citation12 and T7Citation13, that incompletely packaged DNA sometimes has quantized lengths separated by more than the length of a power stroke.
Thus, an author (PS) proposed that a phage DNA packaging motor has two cycles: a packaging ATPase-derived (type 1) cycle and a back-up (type 2) cycle derived from shell expansion/contraction that is sometimes quantized. The proposed type 2 cycle expels non-DNA molecules (mostly RNAs and proteins) that have been accidentally packaged and have stalled the type 1 cycle (type 2 cycle hypothesis).Citation14
The type 2 cycle hypothesis predicts that increasing the host cell's protein and RNA concentration will divert capsid II () to states of both hyper-expansion and contraction during infection with T3. The reason is increased type 2 cycle activation caused by increased accidental host RNA/protein packaging caused, in turn, by increased RNA/protein concentration.
We tested this prediction by, first, adapting phage T3 to propagation in 0.94 M sucrose-supplemented medium, at 30 °C. The medium was modified 2xLB: 0.1 M NaCl, 2% tryptone, 2% yeast extract, 0.08% low-melt Lonza agarose. The sucrose generated osmotic pressure of 27 atmospheresCitation15 The host was Escherichia coli BB/1. The sucrose shrank E. coliCitation16 (The normal internal osmotic pressure of E. coli B is over 3x lowerCitation17) and increased intracellular concentration of osmolytes, RNAs and proteins.Citation18 This T3 adaptation produced three mutations, based on outsourced whole genome Illumina sequencing,Citation19 with sequence changes determined via breseq software20: (1) gene 2.5 single-stranded DNA binding protein, A16T (GCT: ACT); (2) gene 12 tail protein, T514A (ACT: GCT) and (3) gene 16 internal core stack/cylinder protein, P613L (CCA: CTA). The mutant, called T3Su-1, had latent period of 3.5–4.0 hr in 0.94 M sucrose-supplemented medium and 1.1–1.3 hr in medium without sucrose. The latent period of wild type T3 was 0.45–0.55 hr in medium without sucrose.
Next, we produced a lysate by infection with a frozen/thawed T3Su-1 plaque. At the time of infection, log phase host E. coli cells (1.4 × 108/ml) were in a 1.0-liter liquid culture in 0.94 M sucrose-supplemented modified 2xLB medium. We incubated with aeration for 25.5 h at 30 °C. We then purified (1) T3Su-1 capsid I (procapsid; ), (2) DNA-detached capsid II, and (3) T3Su-1 phage, by centrifugation in cesium chloride density gradients.,Citation19,21 Most wild type capsid II has previously been found to have detached from DNA, post lysis. DNA-detached capsid II was produced with kinetics of a phage precursor.Citation22
T3Su-1 phages progressively lost infectivity, which is why a frozen plaque was used for inoculation above. Per ml of lysate, 1.0 × 1010 phage particles were produced, 1% infective, in contrast to 30–40% infective wild type phage. T3Su-1 phage particles were quantified by optical density at 260 nmCitation23 after purification in cesium chloride density gradients.Citation19,21 The T3Su-1 capsid I had the radius of wild type capsid I ± 4%, as determined by native gel electrophoretic sieving (technique of reference Citation19; data not shown). We have assumed here that the T3Su-1 capsid II shell had capsid I stoichiometry, as foundCitation24 by cryo-EM for wild type T7 and as supported below.
Finally, we found that, like wild type T3 capsid IICitation22 T3Su-1 capsid II formed both low (NLD)- and high (NHD)-density bands, during buoyant density centrifugation in a density gradient of either Metrizamide (molecular weight, 789) or its commercial replacement, Nycodenz (molecular weight, 821). The low density of wild type NLD capsid II was caused by impermeability to Nycodenz/Metrizamide.Citation22 This impermeability implied no hole larger than 1.2 nm (intact shell). However, as seen via light scattering, the T3Su-1 low-density capsid II band was not at the position of the band of wild type NLD capsid II, 1.086 g/ml. Instead, T3Su-1 low-density capsid II formed a band of floating particles at the top of the density gradient, implying density of either 1.06 g/ml or lower. Visually, the band formed was granular, with slightly green tint (not shown). The granularity suggested aggregation, which explained the green tint via Mie scattering.Citation25 The result with wild type NLD capsid II has obtained over 10 times; the result with T3Su-1 capsid II has been obtained 4 times.
The protein composition was that of wild type NLD capsid II for both the floating T3Su-1 capsid II (to be called ultra-or U-NLD capsid II) and particles in neighboring fractions of the Nycodenz density gradient (; numbers above lanes indicate densities in g/ml). No host proteins were observed. Host outer membrane vesicles, for example, have higher densities.
The low density of T3Su-1 U-NLD capsid II could only have been caused by water-association, > 3.7 g water per g protein (equations used,Citation22,26). However, proteins have not been found to have bound-water hydrations this high. Thus, as for NLD capsid II, the high hydration of U-NLD capsid II was caused by impermeability to Nycodenz. Importantly, when the total water volume was calculated (equationsCitation22,26), the average U-NLD capsid II particle was larger by at least 8% than NLD capsid II (hyper-expanded U-NLD capsid II).
Existence of hyper-expansion explained the unusually large apparent size of some U-NLD capsid II, when observed by electron microscopy (EM). Specimens were negatively stainedCitation27,28 with 1% sodium phosphotungstate, pH 8.6. Most capsids were, as expected, in aggregates (); most fields were empty. Both aggregated particles and particles at aggregate edges (higher magnification of the latter: ) included some appearing larger than (1) tail-free T3 phage (headsCitation19), observed in a separate specimen (; same magnification) and (2) rare (< 1% of the total) conventionally appearing capsid II (C-CII), observed in the same specimen (arrows in ).
Figure 2. EM at higher magnification of (A–C) T3Su-1 U-NLD capsid II, (D) T3 heads from the study of reference Citation19, and (E) T3Su-1 NHD capsid II.
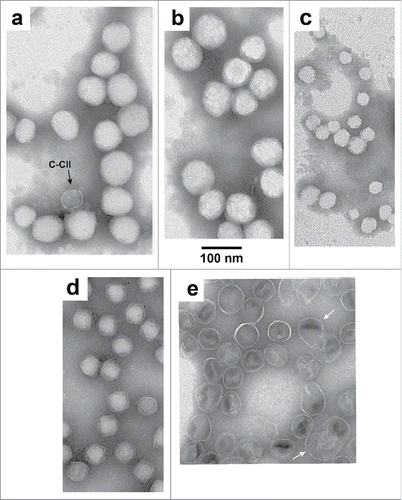
This apparent hyper-expansion was real, based on the following. In a previous studyCitation27 negatively stained (dehydrated) phage T7 particles appeared smaller than hydrated T7, unless flattened. Flattening can increase apparent radius up to 21/2 times. The radius of dehydrated, unflattened T7 (and presumably T3) was 22–23 nm.Citation27 Thus, the largest apparent radius possible for negatively stained U-NLD capsid II was 33 ± 1 nm. This radius was significantly smaller than the radius observed for the larger U-NLD capsid II particles, 35–50 nm. This implies hyper-expansion of the latter, constituting ∼30% of the total; the others are discussed below. In addition, some of the larger-than-35 nm particles were not completely flattened. The reason is that they had structure that, because of its irregularity, was from precipitated, internal buffer components, not the shell ().
Even though U-NLD capsid II particles had no DNA detected in a previous preparative cesium chloride density gradient (< 5% of a genome), almost all (> 99%) had electron density less than observed for heads during EM. The heads were full of negative stain-blocking DNA.Citation19 T3Su-1 NHD capsid II had higher electron density than heads (). We concluded that the low electron density of T3Su-1 U-NLD capsid II was from impermeability to the staining anion, phosphotungstate, which is larger than a Nycodenz molecule.
Hyper-expanded U-NLD capsid II was accompanied by additional low electron density-particles that were contracted, based an apparent radius smaller than 22–23 nm (∼35% of the total). The contracted NLD capsid II particles were seen in the aggregates (). They were also seen un-aggregated, generally in clusters (). Contracted NLD capsid II particles might have been present at lysis, acquiring ultra-low Nycodenz-density via aggregation with hyper-expanded U-NLD capsid II. Alternatively, these particles might have been conversion products of hyper-expanded U-NLD capsid II, the conversion possibly occurring during specimen preparation for EM.
The radii of some U-NLD capsid II particles were not extreme enough to be certain of hyper-expansion or contraction. We speculate that all low electron density-particles were either hyper-expanded or contracted.
In any case, the prediction made above was found to be correct. Wild type NLD capsid II, a form of C-CII, has been at least partially diverted to hyper-expanded U-NLD capsid II. Contracted capsid II has also been seen for the first time, although possibly a conversion product of the hyper-expanded version.
Nonetheless, one reasonably asks whether U-NLD capsid II particles were on-pathway to infective phage. If they were, the correct prediction becomes strong support for the type 2 cycle hypothesis. However, these particles might also have been abortive end products. On-pathway status was suggested by the observed sealing without > 1.2 nm holes. Abortive end products of capsid assembly, such as those for T728, always have had outer shells with holes > 1.2 nm. Similarly, complexity of shell subunit interactionsCitation24 implies that, during DNA packaging, the shell probably doesn't (abortively) slip into a well-sealed structure. Each subunit has several stereo-specific interactions with neighbors, including non-covalent topological linking.Citation24 Thus, the sealing implied by the ultra-low density suggests selection for function in DNA packaging. Similarly, liquid water sealing of bacterial and algal gas vesicles has a selected function, flotation.Citation29,30
Sealing also made unlikely that radius variability was from stoichiometry that varied via subunit exchange among capsids. This exchange would require scaffolding protein-deficient, gap-free shell assembly, which has never been observed. Finally, the shell of U-NLD capsid II must have thinned from 2.0 nm to 1.0 nm or less to cover the surface during hyper-expansion. Formation of β-sheet structure is one way to produce a 0.7–1.0 thick shell.Citation31 We speculate that core stack protein, gp16, was involved in radius alteration/shell thinning because it was mutated in T3Su-1 (above) and can alter shell assembly.Citation28
In summary, the type 2 cycle hypothesis passed its first test. More tests are in progress. This first test depended on directed evolution-based genetics to detect previously obscure states of capsid II. This strategy departed from the previous strategy (recent reviewCitation32) of using conditional lethal mutation-based genetics to block specific stages in phage assembly. That is to say, we introduced a paradigm shift in genetics-based analysis of phage assembly.
Abbreviations
| C-CII | = | conventionally appearing capsid II |
| EM | = | electron microscopy |
| NHD | = | Nycodenz high density |
| NLD | = | Nycodenz low density |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. Wen Jiang, Purdue University, for comments on a preliminary draft of this manuscript. We thank Dr. James J. Bull for providing access to the facility used for sequencing DNA, at The University of Texas at Austin. We thank Drs. Jeffrey Barrick and Borries Demeler for assistance with downloading and running breseq software.
Funding
This work was supported by the Welch Foundation (AQ-764). The Welch Foundation did not participate in the research design, writing and submission of this manuscript.
References
- Liu S, Chistol G, Bustamante C. Mechanical operation and intersubunit coordination of ring-shaped molecular motors: insights from single-molecule studies. Biophys J 2014; 25:1844-58; http://dx.doi.org/10.1016/j.bpj.2014.03.029
- Howard J. Motor proteins as nanomachines: The roles of thermal fluctuations in generating force and motion. Séminaire Poincaré 2009; VII:33-44.
- Kim H, Ha T. Single molecule nanometry for biological physics. Rep Prog Phys 2013; 76:016601; PMID:23249673; http://dx.doi.org/10.1088/0034-4885/76/1/016601
- Serwer P, Jiang W. Dualities in the analysis of phage DNA packaging motors. Bacteriophage 2012; 2:1-17; PMID:22666651; http://dx.doi.org/10.4161/bact.23829
- Liu S, Chistol G, Hetherington CL, Tafoya S, Aathavan K, Schnitzbauer J, Grimes S, Jardine P, Bustamante C. A viral packaging motor varies its DNA rotation and step size to preserve subunit coordination as the capsid fills. Cell 2014; 157:702-13; PMID:24766813; http://dx.doi.org/10.1016/j.cell.2014.02.034
- Cao S, Saha M, Zhao W, Jardine PJ, Zhang W, Grimes S, Morais MC. Insights into the structure and assembly of the bacteriophage phi29 double-stranded DNA packaging motor. J Virol 2014; 88:3986-96; PMID:24403593; http://dx.doi.org/10.1128/JVI.03203-13
- Serwer P. The XXIIIrd phage/virus assembly meeting. Bacteriophage 2014; 4:e27272: Citation of the abstract of Dai et al; PMID:24498537; http://dx.doi.org/10.4161/bact.27272
- Casjens SR. The DNA-packaging nanomotor of tailed bacteriophages. Nature Rev Microbiol 2011; 9:647-57; http://dx.doi.org/10.1038/nrmicro2632
- Rao VB, Feiss M. Mechanisms of DNA packaging by large double-stranded DNA viruses. Annu Rev Virol 2015; 2:351-78; PMID:26958920; http://dx.doi.org/10.1146/annurev-virology-100114-055212
- Black LW. Old, new, and widely true: The bacteriophage T4 DNA packaging mechanism. Virology 2015; 479-80:650-6; PMID:25728298; http://dx.doi.org/10.1016/j.virol.2015.01.015
- Bjornsti MA, Reilly BE, Anderson DL. Morphogenesis of bacteriophage phi29 of Bacillus subtilis: oriented and quantized in vitro packaging of DNA protein gp3. J Virol 1983; 45:383-96; PMID:6185695
- Fang PA, Wright ET, Weintraub ST, Hakala K, Wu W, Serwer P, Jiang W. Visualization of bacteriophage T3 capsids with DNA incompletely packaged in vivo. J Mol Biol 2008; 384:1384-99; PMID:18952096; http://dx.doi.org/10.1016/j.jmb.2008.10.012
- Khan SA, Hayes SJ, Watson RH, Serwer P. Specific nonproductive cleavage of packaged bacteriophage T7 DNA in vivo. Virology 1995; 210:409-20; PMID:7618276; http://dx.doi.org/10.1006/viro.1995.1357
- Serwer P. Proposed Ancestors of Phage Nucleic Acid Packaging Motors (and Cells). Viruses 2011; 3:1249-80; PMID:21994778; http://dx.doi.org/10.3390/v3071249
- Frazer JCW, Myrick RT. The osmotic pressure of sucrose solutions at 30° C. J Am Chem Soc 1916; 38:1907-22; http://dx.doi.org/10.1021/ja02267a001
- Koch AL. Shrinkage of growing Escherichia coli cells by osmotic challenge. J Bact 1984; 159:919-24; PMID:6384186
- Scheie P. Osmotic pressure in Escherichia coli as rendered detectable by lysozyme attack. J Bact 1973; 114:549-55; PMID:4574692
- Cayley S, Record MT, Jr. Large changes in cytoplasmic biopolymer concentration with osmolality indicate that macromolecular crowding may regulate protein-DNA interactions and growth rate in osmotically stressed Escherichia coli K-12. J Mol Recognit 2004; 17:488-96; PMID:15362109; http://dx.doi.org/10.1002/jmr.695
- Serwer P, Wright ET, Liu Z, Jiang W. Length quantization of DNA partially expelled from heads of a bacteriophage T3 mutant. Virology 2014; 456-7:157-70; PMID:24889235; http://dx.doi.org/10.1016/j.virol.2014.03.016
- Deatherage DE, Barrick JE. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Meth Mol Biol 2014; 1151:165-88; http://dx.doi.org/10.1007/978-1-4939-0554-6_12
- Serwer P, Wright ET, Chang JT, Liu X. Enhancing and initiating phage-based therapies. Bacteriophage 2014; 4:e961869; PMID:26713220; http://dx.doi.org/10.4161/21597073.2014.961869
- Serwer P. A metrizamide-impermeable capsid in the DNA packaging pathway of bacteriophage T7. J Mol Biol 1980; 138:65-91; PMID:7411607; http://dx.doi.org/10.1016/S0022-2836(80)80005-2
- Bancroft FC, Freifelder D. Molecular weights of coliphages and coliphage DNA. I. Measurement of the molecular weight of bacteriophage T7 by high-speed equilibrium centrifugation. J Mol Biol 1970; 54:537-546; PMID:4923668; http://dx.doi.org/10.1016/0022-2836(70)90124-5
- Guo F, Liu Z, Fang PA, Zhang Q, Wright ET, Wu W, Zhang C, Vago F, Ren Y, Jakana J, Chiu W, Serwer P, Jiang W. Capsid expansion mechanism of bacteriophage T7 revealed by multistate atomic models derived from cryo-EM reconstructions. Proc Natl Acad Sci USA 2014; 111:E4606-14; PMID:25313071; http://dx.doi.org/10.1073/pnas.1407020111
- Ye QL, Yoshikawa H, Bandow S, Awaga K. Green magnetite (Fe3O4): Unusual optical Mie scattering and magnetic isotropy of submicron-size hollow spheres. Appl Phys Lett 2009; 94:063114; http://dx.doi.org/10.1063/1.3079407
- Serwer P. Buoyant density centrifugation of macromolecules in sodium iothalamate density gradients. J Mol Biol 1974; 92:433-48; PMID:167175; http://dx.doi.org/10.1016/0022-2836(75)90290-9
- Serwer P. Flattening and shrinkage of bacteriophage T7 after preparation for electron microscopy by negative staining. J Ultrastruct Res 1977; 58:235-43; PMID:66322; http://dx.doi.org/10.1016/S0022-5320(77)90015-6
- Serwer P. Fibrous projections from the core of a bacteriophage T7 procapsid. J Supramol Struct 1979; 11:321-6; PMID:544920; http://dx.doi.org/10.1002/jss.400110307
- Walsby AE. Gas vesicles. Microbiol Rev 1994; 58:94-144; PMID:8177173
- Tashiro Y, Monson RE, Ramsay JP, Salmond JPC. Molecular genetic and physical analysis of gas vesicles in buoyant enterobacteria. Environ Microbiol 2016; 18:1264-76; PMID:26743231; http://dx.doi.org/10.1111/1462-2920.13203
- Aggelli A, Bell M, Boden N, Keen JN, Knowles PF, McLeish TCB, Pitkeathly M, Radford SE. Responsive gels formed by the spontaneous self-assembly of peptides into polymeric β-sheet tapes. Nature 1997; 386:259-62; PMID:9069283; http://dx.doi.org/10.1038/386259a0
- Aksyuk AA, Rossmann MG. Bacteriophage assembly. Viruses 2011; 3:172-203; PMID:21994726; http://dx.doi.org/10.3390/v3030172

