ABSTRACT
Curli are a type of proteinaceous cell surface filament produced by enteric bacteria such as Escherichia and Salmonella that facilitate cell adhesion and invasion, bio-film formation, and environmental persistence. Curli assembly involves 6 proteins encoded by the curli specific genes A, B, C, E, F, and G. Although CsgA is the major structural component of curli, CsgE, and CsgF, are thought to play important chaperone like functions in the assembly of CsgA into curli. Given that some proteins with chaperone like function have been observed to contain disordered regions, sequence analysis and circular dichroism spectroscopy was used to investigate the possibility that structures of CsgE and CsgF were also disordered. Sequence analysis based on charge and hydrophobicity, as well as using the disorder prediction software PONDR, indicates that both proteins have significant regions of disorder. The secondary structure and unfolding, of CsgE and CsgF, analyzed using circular dichroism spectroscopy suggests that both proteins lack a well defined and stable structure. These observations support the hypothesis that the curli assembly proteins CsgE and CsgF are disordered proteins containing intrinsically disordered regions.
Background and significance
Bacteria produce a variety of proteinaceous cell surface fimbriae that serve as adhesins, mediating the interaction between bacteria and host cells.Citation1,2 Curli, produced by enteric bacteria such as Escherichia and Salmonella, are a class of highly aggregative fimbriae that are involved in cell adhesion and invasion, bio-film formation, and environmental persistenceCitation3-8 Curli assembly is mediated by proteins encoded by the curli specific genes (csg) A, B, C, D, E, F, and G.Citation9 CsgA is the main proteinaceous structural component of curli while CsgB is thought to be the minor component that helps nucleate and assemble CsgA into curli. Although the details of the assembly process are still under investigation, curli formation is thought to occur in the following manner: proteins required for curli assembly are transported across the inner membrane in a Sec-dependant manner; CsgC, CsgE, and CsgF help transport CsgA and CsgB through the periplasm to the outer membrane; at the outer membrane CsgA and CsgB are secreted to the cell surface through the outer membrane lipoprotein channel CsgG; on the cell surface, membrane anchored CsgB, and CsgF, helps nucleate the oligomerization of CsgA into Curli.Citation10-14
While CsgA and CsgB are the structural components, CsgE, CsgF, and CsgG appear to be required for proper curli assembly. Deletion of CsgE was found to impair secretion of CsgA, CsgB, and CsgF to the extra cellular surface, although small amounts of curli like fimbriae were observed.Citation3 CsgE's ability to affect the secretion of CsgA, CsgB, and CsgF appears to involve interaction with the outermembrane lipoprotein CsgG, and it has been suggested that CsgE acts as a gate that prevents the non-specific translocation of material through the CsgG pore.Citation15,16 It has also been postulated that CsgE, together with CsgC, may determine CsgA conformation and its export to the extracellular surface, and thus play a role in curli assembly.Citation14 In the case of CsgF, the absence of the protein was found to cause CsgA to be secreted away from the cell surface and not assemble into curli. CsgF was also found on the extracellular surface, and to also play an important role in the proper localization of CsgB. Although the exact mechanistic role of CsgE, and CsgF in curli assembly is yet to the determined, the general consensus appears to be that these 2 proteins may have chaperone-like protein functions that are required for curli assembly.Citation14,17
Several chaperone proteins, including the periplasmic HdeA which prevents acid induced protein aggregation,Citation18 and Hsp33 which prevents oxidative stress induced protein aggregation,Citation19 have been suggested to contain disordered regions that play an important role in their function.Citation20-22 Therefore, this study sought to investigate the possibility that CsgE and CsgF are intrinsically disordered proteins. Sequence analysis indicates that based on their charge and hydrophobicity both proteins fall near the disorder-order border on a Uversky plot of mean net charge against mean hydrophobicity. Analysis of secondary structure, and protein unfolding, using circular dichroism indicates that the proteins lack a well defined, and stable structure, supporting the hypothesis that CsgE and CsgF may contain intrinsically disordered regions.
Results
Computational disorder analysis of curli related proteins indicates that CsgE and CsgF contain disordered regions. The amino acid sequences of proteins involved in curl formation were analyzed using Uversky plots () of mean net charge against mean hydrophobicity.Citation23 Based on their charge and hydrophobicity CsgC and CsgG fall within the ordered protein region (). In contrast, CsgA, CagB, CsgE, and CsgF have much lower mean scaled hydropathies, and fall near the order-disorder border (). The computer program PONDR-VSL2 was also used to predict disorder within CsgE and CsgF. For CsgE, 20 residues, or approximately 18% of the protein, were predicted to be disordered (, residues highlighted in red). In the case of CsgF a much higher percentage of the protein, approximately 53%, encompassing the N and C-terminal regions of the protein (residues 20 – 59, and 115–138), are predicted to be disordered (, residues highlighted in red). Possible disordered binding regions in CsgF were also predicted using the program ANCHOR, which indicated the presence of 3 possible binding regions, from residues 20 – 26, 74–84, and 98–105.
Figure 1. Computational disorder analysis of curli related proteins. Uversky plots of mean net charge against mean hydrophobicity for proteins involved in curly assembly. (A) CsgA (B) CsgB, (C) CsgC, (D) CsgE, (E) CsgF, and (F) CsgG. Except for CsgC and CsgG, both of which fall within the ordered protein region (black diamond in panels C and F), the values for CsgA, CsgB, CsgE, and CsgF (black diamond in panels A, B, D, and E) fall near the order-disorder boundary (represented by the solid line) suggesting that these could be disordered proteins. The location of a set of ordered (blue), and disordered (red) proteins are shown for comparison (from Ponder.com).
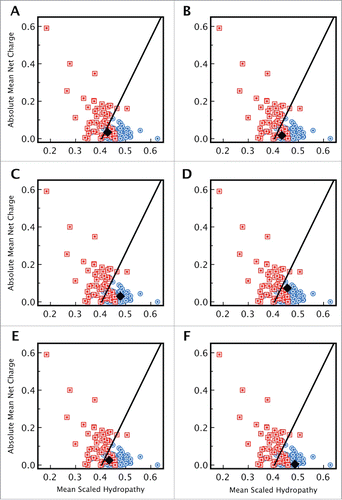
Figure 2. Distribution of disorder and secondary structure within CsgE and CsgF. Sequences of (A) CsgE and (B) CsgF indicating the regions of disorder predicted by PONDR (regions highlighted in red). PSIPred predicted regions of secondary structure are represented using purple rectangles (α-helix) and yellow arrows (β-sheet). Sequence analysis indicates that for CsgE approximately 34% of the residues are in an α-helix, 22% in a β-sheet, and 43% in a random coil, while for CsgF approximately 28 % of the residues are in an α-helix, 27 % are in a β-sheet, with the remainder (45%) being unstructured. The 19 residue signal sequence for each protein is shown in light gray.
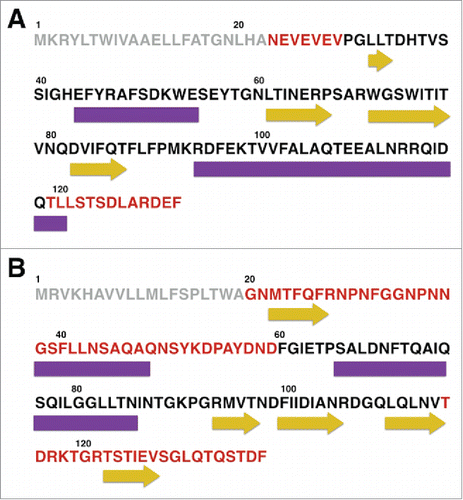
CagE and CsgF contain a significant percentage of unstructured regions. CD spectroscopy was used to investigate the secondary structure of CsgE and CsgF in 20 mM phosphate buffer. CD spectra of both proteins contained a broad negative peak centered at approximately 218 nm (). The broadness of the peak, and the lack of distinct negative minima characteristic of α-helical (208 and 222 nm) or β-sheet (218 nm) structures, is indicative of a structure that contains a mixture of secondary structure elements. In order to quantitate the distribution of secondary structures the online software package DICROWEB was used to deconvolute the experimental CD spectra. Based on this analysis it was estimated that CsgE contains a distribution of 28% α-helical, 23% β-sheet, and 49% random coil structure, while CsgF contains a distribution of 28 % α-helical, 15 % β-sheet, and 57 % random coil structure. The secondary structure of both proteins was also analyzed using PSIPred,Citation24,25 and these estimates agree with the experimental results obtained via CD ().
Figure 3. Circular dichroism spectra of CsgE, and CsgF, and their unfolding with GndHCl. The CD spectra of (A) CsgE and (C) CsgF in solution (black line) are indicative of a mixed distribution of secondary structures, with no distinct characteristic expected for a predominantly α-helical or predominantly β-sheet structure. Spectra were deconvoluted using DICHROWEB and CsgE was estimated to contain a distribution of 28% α-helix, 23% β-sheet and 49% random coil structure. In the case of CsgF the content of α-helix, β-sheet, and coil structure was found to be 28 %, 15 %, and 57 % respectively. The fitted spectra obtained via DICHROWEB are shown in blue. Both (B) CsgE and (D) CsgF exhibited weakly cooperative, or noncooperative unfolding curves with a midpoint denaturation of approximately 2 M GndHCl.
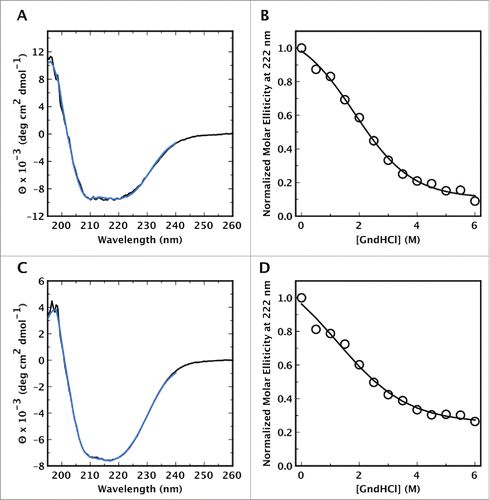
Cage and CsgF undergoes uncooperative unfolding. The stability of CsgE and CsgF was investigated by measuring the GndHCl mediated unfolding of the proteins and monitoring the ellipticity at 222 nm as a function of denaturant concentration (). Both proteins exhibited weakly cooperative unfolding curves with a mid point denaturation at approximately 2.5 M. The lack of cooperativity in the GndHCl induced unfolding transition may be an indication that these proteins lack a well defined, and stable 3-dimensional structure.
Discussion
In the assembly of bacterial curli CsgE and CsgF play important roles with these proteins required for the transport, secretion, and proper assembly of the major curli proteins CsgA and CsgB.Citation3,13,17 The possibility that CsgE and CsgF are disordered proteins (DP) was examined using both computational sequence analysis, and experimental characterization. Based on their charge and hydrophobicity both proteins fall near the disorder-order border on a Uversky plot of mean net charge against mean hydrophobicity ()Citation23 consistent with the possibility that they are DPs. In both proteins the disorder appears to be localized to the N and C-terminal regions (), with a much higher percentage of residues being disordered in CsgF (53%) than in CsgE (20%). In order to experimentally support these predictions the secondary structures of CsgE, and CsgF, were examined using CD spectroscopy. Although both proteins were found to contain a mixture of α-helical and β-sheet elements, a significant portion (approximately 50% for CsgE and 57% for CsgF) of their structures do not appear to have any organized secondary structure. In addition, unfolding curves of the 2 proteins exhibit a gradual loss of structure, characteristic of uncooperative unfolding, rather than the sigmoidal curves obtained during cooperative unfolding. Significant lack of secondary structure, as well as uncooperative unfolding are characteristics of disordered proteins (DPs).Citation22,26-29 Some DPs lack a well-defined tertiary structure over the entire length of the protein, and are known as intrinsically disordered proteins (IDP), while others contain intrinsically disordered regions (IDR) together with regions which are more ordered. Taken together, these CD and bioinformatics analyses suggests that CsgE and CsgF belong to the latter class of DPs, and contain 2 possible disordered regions, rather than being completely disordered.
Although this study focused on characterizing the structure of CsgE and CsgF, the sequences of Csg A,B, C, and CsgG were also analyzed for the presence of disorder. Of the 6 proteins analyzed only CsgC and CsgG are predicted to be ordered (), which is in agreement with previous data that have described the high resolution structure of these proteins.Citation16,30 In the case of CsgA, CD spectra (data not shown), as well as previous accounts,Citation3,31 indicate that the protein adopts an almost completely random coil structure in solution in vitro. This experimental observation supports the prediction that CsgA contains significant disorder as indicated by the finding that the protein falls near the disorder-order border in an Uversky plot (). When associated into curli fibers CsgA has been observed to adopt a structure with β-sheet character (data not shown and refs.Citation3,31). Thus, at least in vivo, CsgA undergoes a disordered to order transition. Both CsgE and CsgF are thought to associate with CsgG (as well as binding to CsgA and/or CsgB). It is possible that such interactions will induce order in these proteins. Indeed a recent report has described the possibility that CsgE undergoes a structural rearrangement upon adopting its functional oligomeric form.Citation32
If, as suggested,Citation17 CsgF provides a chaperon-like function during the transport and assembly of CsgA and/or CsgB, then disordered regions in CsgF could provide the protein interaction surfaces necessary to bind its target. Target binding may be mediated by the predicted disordered binding regions, 2 of which are located in, or near, the terminal regions predicted to be disordered. Although the functional target of CsgF has been suggested to be CsgB,Citation17 and a direct interaction between CsgF and CsgA has not been shown, the demonstration that extracellular CsgF is needed for the proper assembly of CsgB and CsgA into curli, and that in the absence of CsgF both CsgA and CsgB are secreted away from the cell surface, supports a possible in vivo interaction between CsgF and CsgA, in addition to the interaction with CsgB.Citation3,17 Additionally, since CsgB and CsgA share some sequence similarity (˜30%), it is conceivable that if CsgF interacts with CsgB then it could also interact with CsgA, at least in vitro.
Whole cell data clearly indicates that CsgE and CsgF play important roles in the assembly of cell surface associated curli, and one function may be to interact with the curli structural proteins to prevent their intracellular aggregation. The computational and experimental data presented here provides an initial characterization of the structures of CsgE and CsgF. It is possible that these observations obtained in vitro, are not fully representative of the in vivo structures of these proteins, and the possibility that both proteins will adopt a more defined structure upon interacting with their binding partners, as has been shown for other disordered proteins,Citation20 cannot be discounted. However, the data presented here provide compelling evidence to advance the hypothesis that CsgE and CsgF are disordered proteins containing intrinsically disordered regions. Future studies, aimed at characterizing the structure of these 2 proteins at higher resolution, and also investigating their interaction with CsgA, should help develop a more refined understanding of CsgE and CsgF's role in curli assembly.
Methods
Materials. pET21 vectors containing the inserted sequences for CsgE and CsgF fused to the plasmid-encoded C-terminal hexahistidine tag were obtained from Genscript. E. Coli BL21(DE) expression competent cells (Cat. # C6010–03), and a C-terminal AntiHis antibody (Cat. # R931–25) were obtained from ThermoFisher Scientific.
Computational analysis of disorder and disordered binding regions. Predictions of disorder were made using PONDR (Molecular Kinetics, Inc., Indianapolis, IN). Predictions of disordered binding regions were carried out using ANCHOR.Citation33
Expression and Purification of C-terminal hexahistidine tagged Salmonella Typhimurium CsgE and CsgF. E. Coli BL21 (DE3) cells, transformed with the appropriate pET21 vector containing the sequence for Salmonella Typhimurium CsgE or CsgF, fused to the plasmid encoded C-terminal hexahistidine tag, were grown, at 37°C, to a OD(595 nm) of between 0.5 – 1. Protein production was induced with the addition of 1 mM IPTG, and the cells harvested by centrifugation after 3 hours of incubation at 37°C. Cells were resuspended in binding buffer (20 mM phosphate, 20 mM imidazole, 500 mM NaCl ) and lysed using a french press (14,000 psi). Unbroken cells, and cell debris, were removed by centrifugation (5000 × g) and histidine tagged protein was recovered from the lysate using 50% (w/v) Ni- resin using the batch mode as described by the manufacturer. After resin bound protein was eluted using elution buffer (20 mM phosphate, 250 mM imidazole, 500 mM NaCl) the eluent was dialyzed against 20 mM phosphate buffer to remove imidazole. The purity of affinity purified histidine tagged CsgF was investigated using SDS PAGE and the presence of the histidine tag was confirmed by western blot using a C-terminal AntiHis antibody.
CD Spectroscopy. CD spectra CsgE and CsgF were obtained using a Jaco 810 spectropolarimeter (Jasco Inc., Easton, MD). Measurements were taken every 0.5 nm at a scan rate of 50 nm/min with an averaging time of 1 s. All spectra were collected between 190 and 260 nm using a 2 mm path length quartz cuvette. Protein concentration was 25 μM. Spectra were corrected by subtracting an appropriate background, and are presented with intensity units of molar elllipticity. Corrected CD spectra were deconvoluted using the online software package DICHROWEBCitation34,35 to obtain the secondary structure content of CsgF.
GndHCl mediated unfolding of CsgE and CsgF. Guanidine Hydrochloride mediated unfolding of CsgE and CsgF was monitored by the decrease in the CD signal at 222 nm as the concentration of denaturant was increased to 6 M. Measurements were done at room temperature in 20 mM phosphate buffer with a protein concentration of 25 μM. After addition of protein, the samples were incubated at room temperature for a minimum of 30 minutes before CD measurements. After subtraction of appropriate background spectra intensity at 222 nm was normalized such that the intensity in the absence of denaturant was 1.
Abbreviations
| CD | = | circular dichroism |
| Csg | = | curli specific gene |
| DP | = | disordered protein |
| GndHCL | = | guanidine hydrochloride |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Author contributions
SAJ conceived and supervised the study, designed the experiments, analyzed data and wrote the manuscript; AG, NP, KO, AA, RC, and SP performed experiments and did initial analysis of data.
Funding
This work was supported by a professional development grant from the California State University, San Marcos to S.A.J.
References
- Fronzes R, Remaut H, Waksman G. Architectures and biogenesis of non-flagellar protein appendages in Gram-negative bacteria. EMBO J 2008; 27:2271-80; PMID:18668121; http://dx.doi.org/10.1038/emboj.2008.155
- Soto GE, Hultgren SJ. Bacterial adhesins: common themes and variations in architecture and assembly. J Bacteriol 1999; 181:1059-71; PMID:9973330
- Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, Hultgren SJ. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 2002; 295:851-5; PMID:11823641; http://dx.doi.org/10.1126/science.1067484
- White AP, Gibson DL, Kim W, Kay WW, Surette MG. Thin aggregative fimbriae and cellulose enhance long-term survival and persistence of Salmonella. J Bacteriol 2006; 188:3219-27; PMID:16621814; http://dx.doi.org/10.1128/JB.188.9.3219-3227.2006
- Sukupolvi S, Lorenz RG, Gordon JI, Bian Z, Pfeifer JD, Normark SJ, Rhen M. Expression of thin aggregative fimbriae promotes interaction of Salmonella typhimurium SR-11 with mouse small intestinal epithelial cells. Infect Immun 1997; 65:5320-5; PMID:9393832
- Cogan TA, Jørgensen F, Lappin-Scott HM, Benson CE, Woodward MJ, Humphrey TJ. Flagella and curli fimbriae are important for the growth of Salmonella enterica serovars in hen eggs. Microbiology (Reading, Engl) 2004; 150:1063-71; PMID:15073315; http://dx.doi.org/10.1099/mic.0.26791-0
- Gophna U, Barlev M, Seijffers R, Oelschlager TA, Hacker J, Ron EZ. Curli fibers mediate internalization of Escherichia coli by eukaryotic cells. Infect Immun 2001; 69:2659-65; PMID:11254632; http://dx.doi.org/10.1128/IAI.69.4.2659-2665.2001
- Barnhart MM, Chapman MR. Curli biogenesis and function. Annu Rev Microbiol 2006; 60:131-47; PMID:16704339; http://dx.doi.org/10.1146/annurev.micro.60.080805.142106
- Hammar M, Arnqvist A, Bian Z, Olsén A, Normark S. Expression of two csg operons is required for production of fibronectin- and congo red-binding curli polymers in Escherichia coli K-12. Mol Microbiol 1995; 18:661-70; PMID:8817489; http://dx.doi.org/10.1111/j.1365-2958.1995.mmi_18040661.x
- Bian Z, Normark S. Nucleator function of CsgB for the assembly of adhesive surface organelles in Escherichia coli. EMBO J 1997; 16:5827-36; PMID:9312041; http://dx.doi.org/10.1093/emboj/16.19.5827
- Hammar M, Bian Z, Normark S. Nucleator-dependent intercellular assembly of adhesive curli organelles in Escherichia coli. Proc Natl Acad Sci USA 1996; 93:6562-6; PMID:8692856; http://dx.doi.org/10.1073/pnas.93.13.6562
- Loferer H, Hammar M, Normark S. Availability of the fibre subunit CsgA and the nucleator protein CsgB during assembly of fibronectin-binding curli is limited by the intracellular concentration of the novel lipoprotein CsgG. Mol Microbiol 1997; 26:11-23; PMID:9383186; http://dx.doi.org/10.1046/j.1365-2958.1997.5231883.x
- Robinson LS, Ashman EM, Hultgren SJ, Chapman MR. Secretion of curli fibre subunits is mediated by the outer membrane-localized CsgG protein. Mol Microbiol 2006; 59:870-81; PMID:16420357; http://dx.doi.org/10.1111/j.1365-2958.2005.04997.x
- Gibson DL, White AP, Rajotte CM, Kay WW. AgfC and AgfE facilitate extracellular thin aggregative fimbriae synthesis in Salmonella enteritidis. Microbiology (Reading, Engl) 2007; 153:1131-40; PMID:17379722; http://dx.doi.org/10.1099/mic.0.2006/000935-0
- Nenninger AA, Robinson LS, Hammer ND, Epstein EA, Badtke MP, Hultgren SJ, Chapman MR. CsgE is a curli secretion specificity factor that prevents amyloid fibre aggregation. Mol Microbiol 2011; 81:486-99; PMID:21645131; http://dx.doi.org/10.1111/j.1365-2958.2011.07706.x
- Goyal P, Krasteva PV, Van Gerven N, Gubellini F, Van den Broeck I, Troupiotis-Tsaïlaki A, Jonckheere W, Péhau-Arnaudet G, Pinkner JS, Chapman MR, et al. Structural and mechanistic insights into the bacterial amyloid secretion channel CsgG. Nature 2014; 516:250-3; PMID:25219853; http://dx.doi.org/10.1038/nature13768
- Nenninger AA, Robinson LS, Hultgren SJ. Localized and efficient curli nucleation requires the chaperone-like amyloid assembly protein CsgF. Proc Natl Acad Sci USA 2009; 106:900-5; PMID:19131513; http://dx.doi.org/10.1073/pnas.0812143106
- Tapley TL, Körner JL, Barge MT, Hupfeld J, Schauerte JA, Gafni A, Jakob U, Bardwell JCA. Structural plasticity of an acid-activated chaperone allows promiscuous substrate binding. Proc Natl Acad Sci USA 2009; 106:5557-62; PMID: 19321422; http://dx.doi.org/10.1073/pnas.0811811106
- Cremers CM, Reichmann D, Hausmann J, Ilbert M, Jakob U. Unfolding of metastable linker region is at the core of Hsp33 activation as a redox-regulated chaperone. J Biol Chem 2010; 285:11243-51; PMID:20139072; http://dx.doi.org/10.1074/jbc.M109.084350
- Bardwell JCA, Jakob U. Conditional disorder in chaperone action. Trends Biochem Sci 2012; 37:517-25; PMID: 23018052; http://dx.doi.org/10.1016/j.tibs.2012.08.006
- Kovacs D, Tompa P. Diverse functional manifestations of intrinsic structural disorder in molecular chaperones. Biochem Soc Trans 2012; 40:963-8; PMID:22988848; http://dx.doi.org/10.1042/BST20120108
- Pavlović-Lažetić GM, Mitić NS, Kovačević JJ, Obradović Z, Malkov SN, Beljanski MV. Bioinformatics analysis of disordered proteins in prokaryotes. BMC Bioinformatics 2011; 12:66; http://dx.doi.org/10.1186/1471-2105-12-66
- Uversky VN, Gillespie JR, Fink AL. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins 2000; 41:415-27; PMID:11025552; http://dx.doi.org/10.1002/1097-0134(20001115)41:3%3c415::AID-PROT130%3e3.0.CO;2-7
- Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 1999; 292:195-202; PMID:10493868; http://dx.doi.org/10.1006/jmbi.1999.3091
- Buchan DWA, Minneci F, Nugent TCO, Bryson K, Jones DT. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res 2013; 41:W349-57; PMID:23748958; http://dx.doi.org/10.1093/nar/gkt381
- Fink AL. Natively unfolded proteins. Curr Opin Struct Biol 2005; 15:35-41; PMID:15718131; http://dx.doi.org/10.1016/j.sbi.2005.01.002
- Babu MM, van der Lee R, de Groot NS, Gsponer J. Intrinsically disordered proteins: regulation and disease. Curr Opin Struct Biol 2011; 21:432-40; PMID:21514144; http://dx.doi.org/10.1016/j.sbi.2011.03.011
- Uversky VN. Unusual biophysics of intrinsically disordered proteins. Biochim Biophys Acta 2013; 1834:932-51; PMID:23269364; http://dx.doi.org/10.1016/j.bbapap.2012.12.008
- Tompa P, Fuxreiter M, Oldfield CJ, Simon I, Dunker AK, Uversky VN. Close encounters of the third kind: disordered domains and the interactions of proteins. Bioessays 2009; 31:328-35; PMID:19260013; http://dx.doi.org/10.1002/bies.200800151
- Taylor JD, Zhou Y, Salgado PS, Patwardhan A, McGuffie M, Pape T, Grabe G, Ashman E, Constable SC, Simpson PJ, et al. Atomic resolution insights into curli fiber biogenesis. Structure 2011; 19:1307-16; PMID:21893289; http://dx.doi.org/10.1016/j.str.2011.05.015
- Wang X, Smith DR, Jones JW, Chapman MR. In vitro polymerization of a functional Escherichia coli amyloid protein. J Biol Chem 2007; 282:3713-9; PMID:17164238; http://dx.doi.org/10.1074/jbc.M609228200
- Wang H, Shu Q, Rempel DL, Frieden C, Gross ML. Continuous and Pulsed Hydrogen-Deuterium Exchange and Mass Spectrometry Characterize CsgE Oligomerization. Biochemistry 2015; 54:6475-81; PMID:26418947; http://dx.doi.org/10.1021/acs.biochem.5b00871
- Mészáros B, Simon I, Dosztányi Z. Prediction of Protein Binding Regions in Disordered Proteins. PLoS Comp Biol 2009; 5:e1000376; http://dx.doi.org/10.1371/journal.pcbi.1000376
- Whitmore L, Wallace BA. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res 2004; 32:W668-73; PMID:15215473; http://dx.doi.org/10.1093/nar/gkh371
- Whitmore L, Wallace BA. Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers 2008; 89:392-400; PMID:17896349; http://dx.doi.org/10.1002/bip.20853

