ABSTRACT
Nonsense-mediated mRNA decay (NMD) couples protein synthesis to mRNA turnover. It eliminates defective transcripts and controls the abundance of certain normal mRNAs. Our study establishes a connection between NMD and the translation factor eIF5A (eukaryotic initiation factor 5A) in human cells. eIF5A modulates the synthesis of groups of proteins (the eIF5A regulon), and undergoes a distinctive two-step post-translational modification (hypusination) catalyzed by deoxyhypusine synthase and deoxyhypusine hydroxylase. We show that expression of NMD-susceptible constructs was increased by depletion of the major eIF5A isoform, eIF5A1. NMD was also attenuated when hypusination was inhibited by RNA interference with either of the two eIF5A modifying enzymes, or by treatment with the drugs ciclopirox or deferiprone which inhibit deoxyhypusine hydroxylase. Transcriptome analysis by RNA-Seq identified human genes whose expression is coordinately regulated by eIF5A1, its modifying enzymes, and the pivotal NMD factor, Upf1. Transcripts encoding components of the translation system were highly represented, including some encoding ribosomal proteins controlled by alternative splicing coupled to NMD (AS-NMD). Our findings extend and strengthen the association of eIF5A with NMD, previously inferred in yeast, and show that hypusination is important for this function of human eIF5A. In addition, they advance drug-mediated NMD suppression as a therapeutic opportunity for nonsense-associated diseases. We propose that regulation of mRNA stability contributes to eIF5A's role in selective gene expression.
Introduction
Protein synthesis is regulated at multiple levels.Citation1 Most translational control mechanisms do not directly influence mRNA stability, but some couple the translation of individual mRNAs to their destabilization, effectively sifting out defective or surplus mRNAs. The most intensively studied of these mechanisms is nonsense-mediated mRNA decay (NMD). NMD deals with mRNAs that have a premature, in-frame, stop (termination or nonsense) codon, whereas no-go decay and non-stop decay dispose of mRNAs with elongation or termination defects, thereby releasing stalled ribosomes from the template.Citation2
NMD was originally recognized as an error-surveillance process, detecting and eliminating aberrant mRNA templates that, if translated, could give rise to C-terminally truncated proteins that are non-functional or deleterious, possibly by acting as dominant-negative inhibitors.Citation3-7 Such NMD-susceptible mRNAs may result from mutations or from errors in transcription, RNA processing or RNA editing that generate a premature termination codon (PTC). PTCs can arise from an mRNA sequence change that directly creates a nonsense codon, or from an insertion or deletion event that leads to a stop codon. In addition, NMD can be triggered by alternative translation events, such as frameshifting or failure to incorporate selenocysteine, that lead to premature termination.
Beyond its protective role, geared to clearing the mRNA pool of faulty transcripts, NMD regulates gene expression through the programmed destabilization of selected mRNA isoforms.Citation4,8-10 Generation of one such class of substrates entails alternative mRNA splicing, which produces an isoform that is sensitive to NMD and is thereby eliminated, as well as a functional isoform that is translated into protein product. Examples include mRNA isoforms produced by exon inclusion or skipping, or containing an extended exon resulting from the use of alternative 5′ or 3′ splice sites. This regulatory mechanism is termed alternative splicing coupled to NMD (AS-NMD) or regulated unproductive splicing and translation (RUST).Citation11,12
NMD participates in physiological processes such as development and differentiation,Citation4,8 and commands clinical attention because of its role in a large set of genetic diseases referred to as nonsense-associated diseases (NADs).Citation13-15 Well-known NADs include subtypes of β-thalassemia, Duchenne muscular dystrophy and cystic fibrosis; others, though often individually rare, are numerous and prevalent in the population. It has been estimated that PTCs are responsible for one-third of genetic disorders and many types of cancer,Citation16 and that nonsense mutations account for ∼11.5% of human inherited disease.Citation17 Of these single-nucleotide mutations, the majority (∼86%) were predicted to be susceptible to NMD, which has led to ongoing efforts to ameliorate disease by suppression therapy, allowing read-through of nonsense codons, or other means.Citation15,18
Discriminating PTCs from authentic termination signals demands sophisticated biochemical machinery. According to prevailing models, PTCs are distinguished from authentic stop codons by mechanisms that recognize features of the mRNA and of its splicing history.Citation3-5,7,19,20 Three ‘up-frameshift’ factors, Upf1, Upf2 and Upf3 (Smg-2, −3 and −4 in Caenorhabditis elegans), are conserved in evolution and play key roles in NMD. In addition, proteins are involved that interact with the mRNA, ribosomes, and one another, including translation termination and release factors, which interact with the stop codon and the ribosome, and components of the exon junction complex (EJC) that remains bound to exon-exon boundaries after mRNA splicing and nucleocytoplasmic transport. These interactions provide a biochemical basis for the observation that NMD in mammalian cells is frequently triggered by a nonsense codon located >50–55 nucleotides (nt) upstream of a splice junction.Citation21 One model envisions that translating ribosomes stall at the PTC, fail to displace downstream EJCs, and then cooperate with them to recruit Upf1 and other constituents of the mRNA decay machinery leading to accelerated turnover.Citation5
eIF5A is an abundant, highly conserved, and essential protein in eukaryotes.Citation22,23 It was originally characterized as a mammalian translation initiation factor promoting the formation of the first peptide bond.Citation24,25 More recently, roles in polypeptide chain elongationCitation26,27 and terminationCitation28,29 have been documented in yeast. Studies in yeast and human cells indicate that eIF5A is involved in selective gene expression.Citation30-35 In mammalian cells eIF5A has been associated with several biological processes including apoptosis,Citation36 cancer,Citation22 cell differentiationCitation37 and proliferation,Citation38 nucleocytoplasmic transport,Citation39 transcription,Citation40,41 and viral replication.Citation42-44 In yeast, eIF5A has been implicated in RNA turnover,Citation31,45,46 protein translocation,Citation47 and maintenance of cell architecture.Citation48,49
Two paralogous isoforms exist: the major isoform, eIF5A1, is present in most human cells and tissues, whereas eIF5A2 is chiefly restricted to malignant cells. Both isoforms are characterized by an apparently unique posttranslational modification, hypusine.Citation50,51 Hypusine is generated in two enzymatic steps ().Citation52 In the first step, deoxyhypusine synthase (DHS) catalyzes the covalent attachment of a 4-aminobutyl moiety from spermidine to the side chain of a specific lysine residue in eIF5A. In the second step, deoxyhypusine hydroxylase (DOHH) hydroxylates the aminobutyl moiety using molecular oxygen. DOHH is inhibited by the drugs ciclopirox (CPX) and deferiprone (DEF), resulting in accumulation of deoxyhypusine-modified eIF5A, which appears to be inactive with respect to most eIF5A functions.Citation22 These drugs and other DOHH inhibitors potently inhibit HIV-1 replication in vitro, leading to the elimination of HIV-infected cells from cell cultures and a reduction in viral load in vivo.Citation40,42,53,54At the molecular level, we showed that eIF5A is involved in gene expression driven by the HIV-1 promoter. Specifically, the initiation of transcription from the HIV-1 LTR is inhibited by CPX or DEF, and by siRNA-mediated interference with eIF5A1 levels or with hypusine formation by DHS and DOHH.Citation40
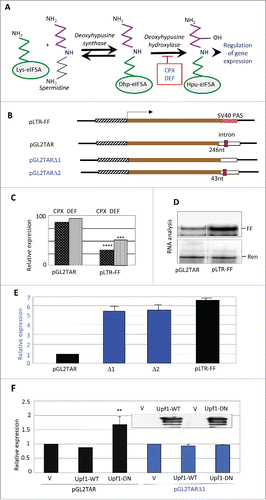
Figure 1. Effects of drugs and eIF5A hypusination on reporter gene expression. (A) Pathway of hypusine formation. The ϵ-amino group of eIF5A Lys-50 (in the human protein) is substituted by a spermidine-derived 4-aminobutyl moiety in a NAD+-dependent reaction catalyzed by DHS. The aminobutyl portion of the deoxyhypusine residue is then hydroxylated by DOHH using O2. DOHH is inhibited by the drugs ciclopirox (CPX) and deferiprone (DEF). (B) Diagrams of plasmids pLTR-FF and pGL2TAR and its deleted derivatives, containing the HIV-1 LTR promoter (hatched), firefly luciferase (FF) coding sequence (brown), and 3′UTRs harboring different SV40 polyadenylation signals (PAS) derived from the vectors pGL2 and pGL3. pLTR-FF contains the SV40 late PAS (blue, stippled) which is devoid of an intron. pGL2TAR contains the SV40 early PAS (open) with a small T intron sequence (red) located 246 nt downstream of the FF luciferase stop codon. In pGL2TARΔ1, the small T intron has been excised. In pGL2TARΔ2, the separation between the FF luciferase stop codon and small T intron sequence is reduced to 43 nt. Arrow: transcription direction. (C) Effect of CPX and DEF on reporter gene expression in 293T cells transfected with pGL2TAR or pLTR-FF. FF activities were normalized to Renilla luciferase expression from the co-transfected pCMV-Ren plasmid and are plotted relative to control cells not treated with drugs. (D) Firefly (FF) and Renilla (Ren) luciferase RNA in 293T cells co-transfected with pGL2TAR or pLTR-FF and pCMV-Ren assayed by RNase protection. (E) Expression from pGL2TAR, pGL2TARΔ1 (Δ1), pGL2TARΔ2 (Δ2), and pLTR-FF in 293T cells. Relative luciferase activity (FF/Ren) is plotted relative to pGL2TAR. (F) Effect of wild-type (WT) or dominant-negative mutant (DN) forms of Upf1 on expression from pGL2TAR and pGL2TARΔ1 in 293T cells. Relative luciferase activity (FF/Ren) is plotted relative to controls transfected with empty vector (V) instead of Upf1 vector. Inset: immunoblot probed for Upf1 protein.
In pursuit of this transcriptional effect, we found that hypusinated eIF5A is functionally associated with NMD in human cells. The relationship was discovered through analysis of reporter gene expression, and was confirmed using a chimeric transcript containing an etiologic PTC that causes β-thalassemia. Transcriptome sequencing identified a set of cellular mRNAs that are coordinately regulated by eIF5A1, DHS, DOHH and Upf1. Prominent in this set are gene transcripts encoding ribosomal proteins, some of which are regulated by AS-NMD. We conclude that eIF5A and its hypusine modification are involved in regulating gene expression at both the protein and mRNA levels, and are possible therapeutic targets in NADs.
Results
Effects of drugs on reporter gene expression
We previously reported that CPX and DEF, which inhibit the second step in the eIF5A modification pathway (), reduce HIV-1 transcription by ∼50% at clinically relevant concentrations.Citation40 Mapping experiments using molecular clones carrying deletions of viral sequences located the drug-responsive element in the viral long terminal repeat (LTR) containing its transcriptional promoter.Citation40 In particular, pLTR-FF, a minimal construct which contains the LTR as the only HIV-derived element driving expression of the firefly luciferase (FF) reporter, was as sensitive to the drugs as the full-length molecular clone.Citation40 In contrast, the drugs elicited little inhibition of firefly luciferase expression in subsequent experiments that used another HIV-1 reporter construct, pGL2TAR (, ). RNA analysis revealed that pGL2TAR generated only ∼20% as much firefly luciferase transcript as pLTR-FF in the absence of drugs (). Because the two constructs have the same promoter and reporter gene, this observation implied a differential effect on RNA decay.
Structural differences between the two constructs led us to postulate that pGL2TAR transcripts, but not those of pLTR-FF, are sensitive to NMD, and that this underlies their discrepant drug sensitivities. pGL2TAR and pLTR-FF are based on the vectors pGL2 and pGL3, respectively, which are related but differ in a number of respects.Citation55 A conspicuous difference lies in their 3′UTRs, which include a polyadenylation signal (PAS). pGL2 contains the SV40 early PAS which has the small T intron sequence upstream, whereas pGL3 contains the late SV40 PAS which does not contain an intron. Thus, pGL2TAR transcripts harbor an intron between the luciferase coding sequence and the poly(A) tail (). Being separated by >50–55 nt from the luciferase stop codon, the intron would be predicted to render pGL2TAR transcripts susceptible to NMD.Citation21 Transcripts of pLTR-FF lack the intron and would be unaffected by NMD, consistent with the ∼5-fold higher level of firefly luciferase mRNA that it produced in comparison with pGL2TAR ().
To test this premise, we generated two deletions in the pGL2TAR construct, each predicted to eliminate its potential for NMD. pGL2TARΔ1 lacks the small T intron, while pGL2TARΔ2 has reduced spacing between the firefly luciferase stop codon and the intron (43 nt instead of 246 nt; ). Both deletions increased firefly luciferase expression to levels similar to that given by pLTR-FF (), consistent with the susceptibility of pGL2TAR to NMD. As an additional test, we transfected cells with a vector encoding a dominant-negative form of Upf1 that suppresses NMD.Citation56 Over-expression of the mutant increased firefly luciferase production from pGL2TAR but not from pLTR-FF, whereas over-expression of wild-type Upf1 had no effect on either of the reporter constructs (). We conclude that pGL2TAR transcripts are sensitive to NMD, unlike those from pLTR-FF.
These findings suggest that CPX and DEF suppress NMD, providing an explanation for the relative insensitivity of pGL2TAR expression to the drugs compared to pLTR-FF (). We propose that the drugs inhibit luciferase transcription from both vectors, but for pGL2TAR the inhibition is counterbalanced by transcript stabilization due to suppression of NMD. In essence, for this construct, drug-induced relief from NMD largely masks the inhibition of transcription from the HIV-1 promoter.
Hypusine-containing eIF5A is involved in mammalian NMD
Since CPX and DEF inhibit hypusination of eIF5A by blocking DOHH, our observations pointed to a connection between human eIF5A and NMD. There is evidence connecting eIF5A to mRNA decayCitation45,46 and NMDCitation31 in yeast, but to our knowledge the relationship has not been studied in higher cells. We therefore examined the effect of inhibiting eIF5A production and modification on NMD in human cells. RNA interference (RNAi) was used to deplete HeLa cells for eIF5A1 (the predominant isoform) or for DHS or DOHH (responsible for modifying eIF5A Lys-50 to deoxyhypusine and then hypusine; ). Upf1, an RNA helicase essential for NMD,Citation56 was silenced for comparison. Expression was measured from pGL2TAR and from a vector encoding a well-characterized NMD substrate, RLuc-Gl Ter,Citation57 which contains a segment of the hemoglobin β-chain harboring a PTC at codon 39 (CAG/glutamine → UAG/stop). This PTC is responsible for a form of βº-thalassemia that predominates in the western Mediterranean basin and peoples originating therefrom. The β-globin segment is fused downstream of the Renilla luciferase gene (), conferring NMD-susceptibility on the chimeric transcript.
Figure 2. Hypusinated eIF5A participates in NMD in human cells. (A) Structure of NMD-sensitive expression construct Rluc-Gl Ter containing the Renilla luciferase gene (RLuc, open rectangle) fused in frame at its 3′ end to three exons of the human β-globin gene (Gl, filled rectangles), the second of which harbors a PTC at position 39. (B, C): Effect of siRNA directed against eIF5A1 (5A), Upf1, DHS or DOHH on expression from pGL2TAR (left) or Rluc-Gl Ter (right) in HeLa cells with co-transfected pCMV-Ren and pCMV-FF, respectively. Relative luciferase activity (FF/Ren, left; Ren/FF, right) is plotted relative to expression transfected with control siRNA (siC). (D) Effect of the drugs DEF and CPX at concentrations indicated on expression from RLuc-Gl Ter. Relative luciferase activity (Ren/FF) is plotted relative to expression in the absence of drug. (E) Effect of drugs on Renilla luciferase RNA measured in Northern blots. Data is mean of three independent experiments presented as a ratio of expression from RLuc-Gl Ter (NMD sensitive) relative to the normal construct, RLuc-Gl Norm. *, p < 0.05.
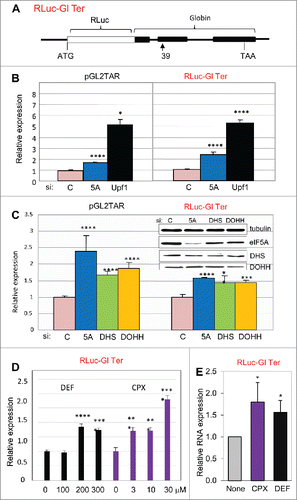
As expected, depletion of Upf1 increased reporter activity from both NMD-sensitive vectors (). eIF5A1 depletion also increased reporter activity (), albeit to a lesser extent. This difference may reflect the exceptional abundance and stability of eIF5A in HeLa cells.Citation58 Luciferase expression from the pGL2TAR and RLuc-Gl Ter reporters was also increased when cells were depleted for DHS or DOHH (). Correspondingly, treatment with DEF and CPX also increased expression from the chimeric NMD substrate RLuc-Gl Ter (). For both drugs, the concentration dependence corresponded to that reported previously for inhibition of eIF5A modificationCitation40; particularly notable is the apparent threshold between 100 μM and 200 μM DEF, observed in a number of assay systems.Citation40,42,54 Increased luciferase activity was mirrored by increased transcript levels (). These results support the conclusion that eIF5A is associated with NMD in human cells and indicate the importance of hypusine in this role.
Cellular gene regulation by hypusine-containing eIF5A
The preceding experiments were carried out with model constructs carrying a reporter gene upstream of either a viral intron or a PTC-containing mutant human gene segment. As NMD regulates the expression of some normal genes, we set out to determine whether eIF5A influences cellular transcript levels. To identify cellular RNAs that are dependent on eIF5A and its hypusine modification, we used deep sequencing (RNA-Seq) to analyze polyA(+) RNA from HeLa cells depleted for eIF5A1, DHS or DOHH ( and Fig S1A). Transcripts dependent on Upf1 were analyzed in parallel. Knockdown was ∼70–80% effective at the RNA level and was specific (Fig. S1B, C). Knockdown of one component was not accompanied by a reduction in the level of the others, indicating that these genes are not coordinately regulated.
Figure 3. Effects of eIF5A and hypusination on the expression of cellular genes. (A) Flowchart of RNA sequencing analysis. (B) Clustering of genes and samples using gene expression changes. RNA expression changes of 8,690 genes that exhibited significant change relative to siC (t-test, p < 0.05 in at least one condition) are shown on a log2 scale with heatmap values displayed below. Hierarchical clustering was applied based on Pearson correlation and the average linkage method. (C) Scatter plots showing two-way correlations between the effects of eIF5A1, DHS and DOHH knockdowns on gene expression. Fold changes (>1.2, Fisher's exact test p < 0.001) are displayed on a log2 scale and Pearson r values are shown.
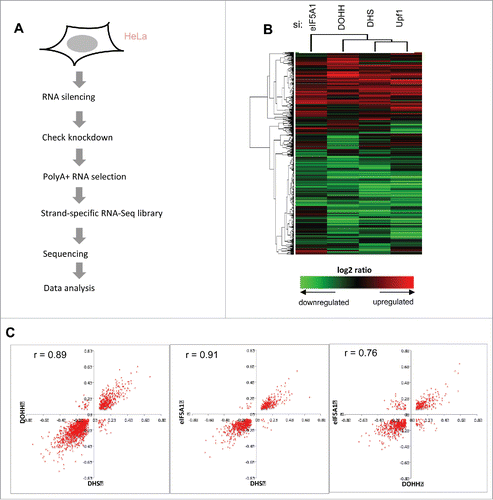
Cluster analysis revealed closely matching patterns of changes in gene expression (). Both increases and decreases in gene expression were observed. Taking a conventional threshold criterion of significance (fold change >1.2, Fisher's exact test p < 0.001), approximately twice as many gene transcripts decreased as increased (∼1.5- to 2.5-fold for individual knockdowns: Fig S2). Pairwise comparisons corroborated the concordant effects of eIF5A, DHS and DOHH knockdowns on the expression of individual genes as illustrated in scatter plots with high correlation coefficients (Pearson's r values) ranging from 0.76 to 0.91 (). The concordance is emphasized by quantitation. For example, among the 469 genes whose expression was up-regulated after eIF5A1 knockdown, almost half (∼45%) were also significantly up-regulated by DHS knockdown; only a few (<1%) were down-regulated, and the remainder did not change significantly. Conversely, among the 719 genes whose expression was down-regulated after eIF5A1 knockdown, ∼64% were down-regulated by DHS knockdown while <1% were up-regulated. Similar response patterns were observed for each pairwise comparison (Fig. S3). Concordant changes outnumbered discordant changes in gene expression by ∼27:1 overall. These observations are as expected for members of a singular pathway and are consistent with the view that DHS and DOHH are dedicated to eIF5A modification.Citation59 If eIF5A1, DHS or DOHH exerts distinct effects on cellular gene expression that are not shared with other pathway members, evidence for such effects was not obvious in this analysis.
Coordinate regulation by Upf1 and hypusinated eIF5A
We examined gene expression under conditions of Upf1 depletion to define the effects of impairing NMD and allow comparison with the effects of obstructing the eIF5A pathway. In accord with the cluster analysis (), Venn diagrams displayed extensive overlaps among the effects of all four RNAi knockdowns (). In pairwise comparisons, Upf1 knockdown showed a marked tendency toward coordinate effects on gene expression with knockdown of eIF5A1, DHS or DOHH. Scatter plots manifested a high degree of correlation (r values 0.86 – 0.93; ), which was confirmed by quantitation (Fig. S4A). The individual correlations were highly significant (p < 0.000005 for eIF5A1, DHS or DOHH vs. Upf1), and the evidence was further strengthened by consideration of transcripts that displayed uniform behavior toward knockdown of all three eIF5A pathway genes. Most (∼73%) of the transcripts that increased with knockdown of the eIF5A pathway also increased when Upf1 was depleted; similarly, most (∼77%) of the transcripts that decreased with knockdown of the eIF5A pathway also decreased when Upf1 was depleted. Nearly all of the remainder did not display significant changes, and only one gene displayed divergent behavior (Fig. S4B, C). In sum, transcripts that were up-regulated by Upf1 depletion were predominantly up-regulated by knockdown of the eIF5A pathway; conversely, transcripts that were down-regulated by Upf1 depletion were overwhelmingly down-regulated by knockdown of the eIF5A pathway.
Figure 4. Coordinate effects of Upf1 and the eIF5A pathway on human gene expression. (A) Venn diagrams displaying the number of genes whose expression changed significantly (fold change >1.2, Fisher's exact test p < 0.001) in HeLa cells depleted for eIF5A1, DHS, DOHH and Upf1 in comparison with siC-treated cells. (B) Scatter plots showing the distribution of genes whose expression was significantly up- or down-regulated by knockdown of Upf1 compared with eIF5A1 (left), DHS (middle) or DOHH (right). Parameters are as in . (C) GO analysis of genes up-regulated by knockdown of Upf1, eIF5A1, DHS or DOHH. The p-values are plotted for each term.
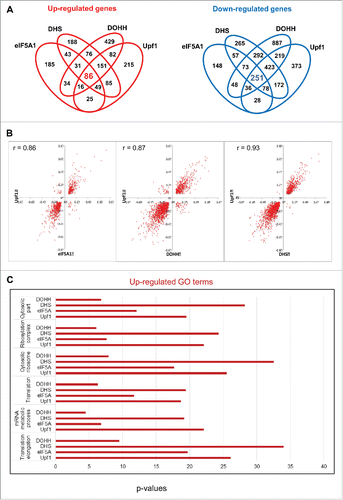
This coordinate response of individual gene transcripts was supported by Gene Ontology (GO) analysis, which indicated a functional relationship between eIF5A and NMD for classes of genes. GO terms that were most enriched with Upf1 depletion were also highly enriched with knockdown of the eIF5A pathway components (Table S1). This co-regulation applied to GO terms that were predominantly up-regulated () and to those that were predominantly down-regulated (Fig. S5). Taken together, the data strongly support the inference that hypusinated eIF5A and Upf1 serve related roles, both positive and negative, in cellular gene expression.
eIF5A-dependent NMD targets in the human transcriptome
Of the 15,634 nuclear gene transcripts identified, 709 (∼4.5%) increased significantly in Upf1 knockdown cells relative to control siRNA-treated cells and are therefore candidates for NMD. The majority of these, 494 (∼70%), also increased significantly in cells knocked down for eIF5A1 and/or its modifying enzymes, and 86 (∼12%) met the most stringent criterion, being significantly increased in cells knocked down for all four gene products (Upf1, eIF5A1, DHS and DOHH; ). These 86 gene transcripts are listed in Table S2 and are considered candidates for eIF5A-associated NMD (5A-NMD).
Figure 5. Up-regulation of cellular transcripts by depletion of Upf1 or eIF5A pathway genes. (A) Gene transcripts significantly up-regulated in RNA-Seq analysis by Upf1 knockdown (total 709) that were also up-regulated by knockdown of eIF5A1, DHS or DOHH singly or in combinations. (B) qPCR analysis showing up-regulation of 10 gene transcripts in cells depleted for eIF5A1, DHS, DOHH or Upf1. Data are normalized to siC-treated cells. Standard deviations are included but are too small to be readily visible; p-values were <0.01, and mostly <0.001, except where marked #. The genes were selected on the basis of their up-regulation in RNA-Seq analysis (see text for details).
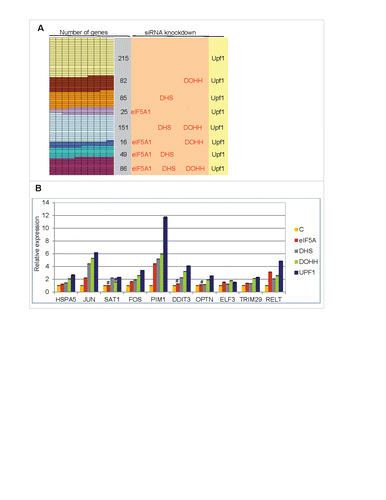
Consistent with a mechanistic commonality, the rank orders of the transcripts were largely coincident based on fold change upon knockdown of Upf1, eIF5A, DHS and DOHH (not shown). Quantitative PCR (qPCR) assays were conducted on 10 selected genes (Table S2 and Fig. S6). Depletion of eIF5A1, DHS, DOHH or Upf1 led to an increase in all RNAs examined (). In most cases, the largest stimulation was seen with Upf1 and the smallest with eIF5A1, presumably reflecting its stability and abundance relative to the other three enzymes (cf. Fig. S1C). These results validate the RNA-Seq data and support the conclusion that the transcripts are regulated by eIF5A-associated NMD.
Regulation of translation system components by eIF5A and AS-NMD
The 5A-NMD set of up-regulated transcripts contains representatives of many gene families, notably including products involved in gene expression (Table S2). The most prominent up-regulated terms in the GO analysis of the knockdown data were “translational elongation” in the Biological Process category and ‘cytosolic ribosome’ in the Cellular Component category (Table S1), with a high level of significance (). Other significantly up-regulated terms included ‘translation’, ‘mRNA metabolic process’, ‘cytosolic part’ and ‘ribonucleoprotein complex’ ( and Table S1). Of the 86 up-regulated 5A-NMD transcripts, 19 (∼22%) encode components of the protein synthesis machinery, including 15 (∼17%) cytoplasmic ribosomal proteins (), corresponding to a >30-fold enrichment compared to their frequency in the database as a whole (∼0.54%). Interestingly, mitochondrial ribosomal protein transcripts were not up-regulated even though they are also transcribed from the nuclear genome.
Table 1. Up-regulated transcripts encoding components of the translation systemFootnote1
For genes in the ‘cytosolic ribosome’ GO category, scatter plots revealed an almost uniform up-regulation (). Among these, transcripts of the L3, L7a and L12 genes (RPL3, RPL7a and RPL12) were significantly up-regulated by knockdown of Upf1, eIF5A1, DHS and DOHH (), while L10a narrowly missed the criteria for inclusion (not shown). Previous studies found that these four cytoplasmic ribosomal proteins are regulated by NMD in C. elegans,Citation12 and that L3 and L12 are NMD-regulated in human cells.Citation60 In addition, mRNAs encoding hnRNP L-like protein (HNRNPLL) and spermidine/spermine N1-acetyltransferase-1 (SAT1 or SSAT) were also up-regulated by knockdown of the eIF5A pathway and of Upf1 (Table S2). HNRNPLLCitation61 and SAT162 transcripts are regulated by NMD in human and mouse cells, respectively, and both are functionally connected to the protein biosynthetic pathway.
Figure 6. Alternative splicing-coupled NMD of ribosomal protein genes. (A) Scatter plot for genes in the “cytosolic ribosome” GO category, displaying co-regulation of transcripts by knockdown of Upf1 and by knockdown of eIF5A1, DHS or DOHH. Parameters are as in (note log scale). (B) Evidence for AS-NMD in RPS3 transcripts. Isoform expression in control and knockdown samples was analyzed by the MISO method, where Ψ represents the fraction of transcripts containing exon 5. Histograms show Ψ value distributions (abscissa, Ψ value; ordinate, percent frequency) with 95% confidence intervals (dotted lines). The Ψ values are listed together with 95% confidence intervals (square brackets). Bayes factors were >1012 in all samples except for the eIF5A1 knockdown (not shown). Diagrams below illustrate the alternatively spliced junctions detected (top) and corresponding RNA isoforms (bottom). Exons and introns are depicted as boxes and lines, respectively; filled and open boxes depict translated and untranslated sequences, respectively.
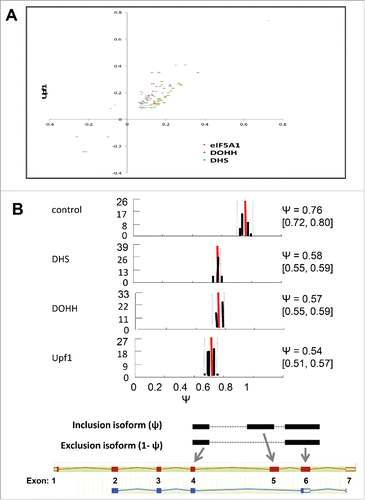
All of these RNAs are regulated by AS-NMD, in which an alternative splicing event results in the introduction of an NMD-determining PTC. This is due to the inclusion of an intron-derived ‘poison exon’ (in SAT1 and HNRPLL) or the extension of an exon at its 5′ end (in RPL3) or 3′ end (in RPL12, RPL7a, and RPL10a) into intronic sequence. Examination of RNA-Seq reads for splice pattern variations revealed changes in the proportions of alternatively spliced mRNAs in several genes that were up-regulated by knockdown of Upf1 and the eIF5A pathway. In the RPS3 and eIF4A2 genes, the knockdowns led to increased accumulation of isoforms that are predicted to be substrates for NMD, implying that RPS3 and eIF4A2 may be subject to regulation by AS-NMD. The proportion of exon 5-skipped RPS3 transcripts increased, and the resulting out-of-frame translation of canonical exon 6 would encounter a PTC 148 nt upstream of the mRNAs 3′-most splice junction (). Similarly, the increased accumulation of an intron-included eIF4A2 gene transcript results in a translational frameshift leading to a PTC that is predicted to cause NMD (data not shown). These findings indicate that hypusinated eIF5A has a role in regulation of gene expression by AS-NMD.
Discussion
This study presents functional and bioinformatic evidence indicating that hypusine-containing eIF5A is important for NMD in human cells. NMD-susceptible transcripts were stabilized when the synthesis or hypusination of eIF5A was compromised by RNAi or drug treatment. Depletion of hypusinated eIF5A stabilized NMD targets including a model substrate with a virus-derived intron downstream of its normal stop codon, and a chimeric transcript containing a pathological PTC responsible for one form of βº-thalassemia. In the human transcriptome, depletion stabilized eIF5A-dependent NMD targets which were especially enriched in components of the translation system and in some cases regulated by alternative splicing. The participation of eIF5A in NMD in human cells is consistent with the broad role of this protein in cell activities, and it may provide a basis for a deeper understanding of its functions and for therapeutic intervention in nonsense-associated genetic diseases. In protein synthesis, its participation in NMD comports with increasing recognition of eIF5A as a regulatory factor involved in several aspects of translation.
Hypusinated eIF5A is functionally associated with NMD
eIF5A and its modifying enzymes, DHS and DOHH, like many components of the NMD machinery, are highly conserved; our findings argue that the functional relationship between them has also been conserved through evolution. Earlier work implicated eIF5A in mRNA decay and NMD in the yeast Saccharomyces cerevisiae. In yeast, several RNAs were stabilized at the non-permissive temperature by mutation of eIF5A,Citation45 a finding that was confirmed with independent eIF5A mutations.Citation46 It is notable that one of the stabilized yeast transcripts was a PTC-containing incompletely spliced pre-mRNA that is an NMD target.Citation45 In a subsequent microarray analysis, several hundred RNAs were found to be up-regulated in yeast harboring mutant human eIF5A1.Citation31 Of these, ∼60% were also up-regulated in cells deficient for Upf genes, including many that appeared to be NMD substrates. Substantial overlaps were observed with the mRNA sets that were up-regulated when other elements of the decay machinery, decapping enzyme and exonuclease, were deleted.Citation31
In our experiments, inactivation of the eIF5A and NMD pathways in human cells also led to increased expression of many transcripts, and similar effects were elicited by depletion of Upf1, eIF5A1, DHS and DOHH. Conspicuous among the up-regulated transcripts were mRNAs encoding components of the translation system, especially cytoplasmic ribosomal proteins (). This finding is consistent with previous observations documenting the regulation of specific ribosomal protein mRNAs by NMD.Citation12,60 In a recent study of EJC distribution on HeLa cell transcripts, exons of ribosomal protein mRNAs were found to be markedly deficient in EJC occupancy,Citation63 leading us to speculate that degradation of at least some ribosomal protein mRNAs (and possibly other 5A-NMD transcripts) may follow an EJC-independent NMD pathway.Citation3,64 Alternative explanations include the possibility that ribosomal protein mRNAs bind EJCs with a different composition than those examinedCitation63 or are more rapidly degraded than other NMD targets when associated with EJCs. Many of the up-regulated transcripts () are abundant, growth-regulated TOP mRNAs. These mRNAs, which bear a 5′ terminal oligopyrimidine (5′ TOP) sequence, are coordinately regulated at the translational level,Citation65,66 and display limited EJC binding.Citation63 Evidently, further analysis will be required to determine the relationships between 5′ TOP sequences, EJC occupancy, eIF5A, and the susceptibility of mRNAs to NMD.
While the impact of hypusination on RNA decay has not been evaluated in yeast, our data show that neither the lysine-containing nor the deoxyhypusine-containing form of eIF5A is sufficient to substitute for the mature hypusinated form of the factor in human cells. This is in keeping with most of the reported functions of human eIF5A, for which hypusine is essential. Exceptionally, deoxyhypusyl-eIF5A is believed to play a role in apoptosis in mammalian cells,Citation36 and it retains some functionality in yeast and in vitro.Citation22,23
Impairment of the eIF5A and NMD pathways also led to decreased expression of some transcripts in human cells, as observed to a lesser extent in yeast.Citation31 The overlap between the two sets of down-regulated transcripts was less pronounced in yeast than in human cells. It remains to be seen whether these differences are due to divergent mechanisms for transcript down-regulation or to technical differences between the yeast and human experiments, but it is noteworthy that the PPP and PPG motifs, a signature of eIF5A dependence, are found less frequently in yeast than human proteins.Citation67
Interplay between eIF5A and NMD
The involvement of eIF5A in NMD suggests a regulatory and possibly proofreading role in translation. While the biochemical basis for the eIF5A-NMD relationship is unknown, in human as well as yeast cells, three general types of explanation can be considered. The most straightforward would be a direct or indirect physical interaction between components of the eIF5A and NMD pathways. eIF5A is exceptionally abundant, even for a protein synthesis initiation factorCitation68,69 and it has been implicated in diverse cellular processes. Accordingly, it is likely to interact with many cellular proteins. On the other hand, NMD is a complex and intricately regulated process entailing many proteins. As well as Upf1–3, these include the nuclear cap-binding proteins CBP80 and CBP20, translation termination and release factors eRF1 and eRF3, the nuclear poly(A)-binding protein N1, components of the EJC, and Smg factors and protein phosphatase PP2A which regulate the phosphorylation status and activity of Upf1. Possibly eIF5A interacts with one or more of these NMD components although preliminary co-immunoprecipitation experiments did not reveal an interaction between Upf1 and eIF5A (M. Hoque, unpublished data). In view of eIF5A's recently demonstrated role in translation terminationCitation28,29 it is particularly tempting to postulate a connection with release factors. Additional potentially relevant interactions are with select mRNAsCitation70 and RNA sequence motifs,Citation71 with ribosomes,Citation72,73 and with numerous cellular proteins, especially ones involved in translation and RNA metabolism and transport, and the cell cycle.Citation74 Conceivably eIF5A's RNA- and/or protein-binding capabilities could regulate mRNA export from the nucleus in such a way as to influence the ‘pioneer round’ of translation that is a key feature of NMD in some models.Citation5 Interestingly, hypusinated eIF5A participates in the hormone-induced degradation of the mRNA encoding the luteinizing hormone receptor (LHR) in rat ovaries. LRBP interacts with both eIF5A and LHR mRNA,Citation75 and RNA turnover appears to be mediated by the LHR mRNA binding protein (LRP) in response to a signaling cascade triggered by human chorionic growth hormone.Citation76
Alternatively, the relationship between eIF5A and NMD could be ascribed to an indirect effect, for example, secondary to the function of eIF5A in translation. Knockdown of eIF5A, or inhibition of its hypusination, would be expected to reduce the synthesis of eIF5A-dependant proteins. Although this class has not been fully defined, one feature in yeast is the presence of oligoproline runs. Similar to its eubacterial ortholog, EF-P, yeast eIF5A was shown to facilitate the decoding of stretches containing PPP or PPG sequences.Citation23 Recent studiesCitation28,29 have expanded the spectrum of eIF5A-dependant sequences considerably, however, and the extent to which this specificity holds for mammalian eIF5A remains to be established. In human cells, a set of proteins has been characterized as eIF5A-dependant, many of which do not share the oligoproline motifs.Citation33,34,67 In bacteria, EF-P may also display a somewhat broader specificity.Citation22 Possibly eIF5A is required for the synthesis of one or more of the proteins involved in NMD. Conversely, the action of Upf1 on RNA stability might affect transcripts of eIF5A pathway genes. One observation that argues against co-regulation of Upf1, eIF5A1, DHS and DOHH, at least in its simplest form, is that their transcripts are not coordinately silenced (Fig. S1C).
A third general explanation takes note of observations that eIF5A affects higher-order functions such as cell structure and behavior. For example, it is implicated in control of cell shape, cell division, growth and cytoskeletal architecture,Citation33,34,49 consistent with down-regulation of GO terms such as ‘cell cycle’ and Golgi organization’ (Table S1 and Fig. S5). Although there is a correlation between oligoproline motif content and actin cytoskeleton-associated function,Citation67 it is uncertain whether these actions are themselves contingent on alterations in gene expression. The abundance of eIF5A lends support to the view that its function is not restricted to protein synthesis, and it is not ruled out that the association of eIF5A with AS-NMD is due to an effect on splicing.
NMD, eIF5A and drugs
Discovery of hypusinated eIF5A's participation in NMD emerged from our analysis of the effects of CPX and DEF on reporter gene expression in human cells. DEF is a metal chelator used clinically to alleviate transfusional iron overload arising in thalassemia, and CPX is a topical antifungal that has recently been administered orally.Citation77 At clinically relevant concentrations, both drugs block DOHH, the enzyme responsible for the second step in this highly specific post-translational modification pathway. Of the transcripts that were uniformly up-regulated by depletion of Upf1, eIF5A, DHS and DOHH, about a third were also up-regulated by the drugs (Fig. S6) and representatives of this subgroup were all confirmed to be up-regulated (), solidifying the conclusion that hypusine-containing eIF5A is implicated in NMD. Remarkably, this subgroup did not include transcripts encoding cytoplasmic ribosomal proteins, even though these were prominently up-regulated by knockdown of Upf1 or the eIF5A pathway. In fact, most cytoplasmic ribosomal protein transcripts were down-regulated by CPX and DEF (data not shown). Mitochondrial ribosomal protein transcripts, which were generally unaffected by knockdown of Upf1 and the eIF5A pathway genes, were also mostly down-regulated by the drugs (data not shown). This may be due to an over-riding negative control at the transcriptional level, reflecting inhibition of cell growth and division exerted by the drugs at the concentrations usedCitation33 as indicated by GO data (Fig. S5).
The involvement of eIF5A in NMD opens a possible route toward treatment of NADs, especially those due to a point mutation such as the β globin mutation tested here (). These account for ∼12% of human genetic diseases and are frequently more severe than those resulting from missense mutations because they lead to the destruction of the aberrant mRNA. One pharmacological strategy to combat NADs would attain production of adequate amounts of full-length protein by (1) suppression of the nonsense mutation, allowing read-through of the PTC by insertion of a functionally acceptable amino acid at the premature stop site; and/or (2) stabilization of the mRNA, by preventing or slowing its decay. Several agents, including aminoglycosides and derivatives, negamycin, and PTC124 (ataluren, Translarna™) have been discovered and developed that can cause read-through.Citation15,18 Some of these have progressed to the clinic with modest but encouraging success. Less attention has been paid to inhibitors of mRNA turnover, however. PI3K inhibitors such as caffeine and wortmannin block Upf1 phosphorylation by Smg1, and NMDI-1 inhibits Upf1 dephosphorylation by blocking its interaction with Smg5.Citation15,18 Amlexanox stabilizes several pathogenic nonsense-containing mRNAs leading to production of full-length protein.Citation78 This drug is an anti-inflammatory, anti-allergic immunomodulator, and inhibits G protein-coupled receptor kinases such as GRK5.Citation79 Although its mechanism of action against RNA decay is not known, it appears to differ from that of CPX and DEF in that it does not stabilize normal cellular transcripts.
Combination of an mRNA stabilizing drug with a read-through agent could be a potent therapeutic strategy widely applicable to a large number of NADs. Moreover, it is conceivable that elevation of mRNA levels alone may be sufficient to achieve therapeutic benefit in some diseases or individuals in the absence of nonsense suppression therapy, or together with other treatment modalities.
The eIF5A regulon
To our knowledge, this report is the first to address the global influence of eIF5A and its hypusine modification on the human transcriptome. eIF5A controls the expression of a suite of human genes, referred to as the eIF5A regulon, through modulation of transcript levels and mRNA translation.Citation33 In HeLa cells we observed the regulation of different categories of genes by CPX and DEF: Group 1 proteins (HSP27, NM23 and DJ-1) were down-regulated at the translational level, whereas Group 2 proteins (TrpRS and PRDX2) were up-regulated at the mRNA level. Presumably at least some of the up-regulated transcripts observed here will prove to be members of Group 2. Proteomic analyses have defined large sets of gene products that are increased or decreased in abundance at the protein level. In pancreatic cancer cells, eIF5A1 depletion up-regulated proteins involved in mRNA and protein production and the stress response, and down-regulated proteins in several GO categories including protein targeting and localization.Citation34 In HeLa cells, there was up-regulation of proteins in the cellular component organization and protein folding categories, and down-regulation in the metabolic processes category, emphasizing effects on ER stress.Citation35 Despite striking similarities, considerable divergence was also evident among these analyses, presumably reflecting differences in cell types, analytical techniques, and other experimental details. In view of the action of eIF5A on both transcription and translation, a comprehensive description of the eIF5A regulon will require coordinate analysis of effects on both mRNA and protein populations. The observation that eIF5A strongly regulates the expression of components of the translation system suggests that the eIF5A operon may act as a switch or rheostat controlling cell growth.
Materials and methods
Cells, cell culture, immunoblotting and luciferase assays
Cells from the American Type Culture Collection were maintained as recommended and seeded one day before treatment. CPX (Sigma-Aldrich) and DEF (Calbiochem) were freshly dissolved in PBS and added to the medium for 24 hr at 30 μM and 250 μM, respectively, except where otherwise noted. Luciferase assays and immunoblotting were conducted as described previously.Citation40 Duplicate luciferase assays were repeated at least twice. The results were normalized as stated in legends, and are displayed together with standard deviations and Student's t-test values relative to respective controls indicated according to the convention: *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p <0.0001.
Plasmids
The HIV-1 reporter plasmids, pGL2TAR and pLTR-FF, and controls pCMV-FF and pCMV-Ren were described previously.Citation40 Deletion constructs pGL2TARΔ1 and pGL2TARΔ2 were generated by replacing the 3′UTR sequences of pGL2TAR with PCR-amplified sequences lacking intron sequences or 203 nt at the 5′ end, respectively. All sequences were confirmed by sequencing. The plasmids FLAG-Upf1WT, FLAG-Upf1DN (FLAG-Upf1R844C) and empty vector,Citation56 and pRLuc-Gl Norm (Gln39) and pRLuc-Gl Ter (PTC at codon 39),Citation57 were generously provided by Dr. Lynne Maquat (University of Rochester).
RNA interference
HeLa cells were transfected with 20 nM siRNA and 293 cells with 50 nM siRNA using Hiperfect (Qiagen). Cells were harvested at 24 hr post-transfection unless otherwise stated. eIF5A1 siRNA was as defined previously.Citation33 DHS, DOHH and Upf1 “ON-TARGET plus SMART pool” siRNAs and non-targeting control siRNA (siC) were from Dharmacon.
RNA analysis
RNase protection assays were conducted as described.Citation40 For northern analysis, HeLa cells transfected with pRLuc-Gl Ter, pRLuc-Gl Norm and pCMV-FF were treated with 30 µM CPX, 300 µM DEF or no drug. Total RNA was isolated 24 h later, blotted, and assayed using radiolabeled antisense probes. RNA bands were quantitated with ImageJ 1.43u software (NIH) and the Renilla luciferase RNA signal was normalized to FF RNA. For qRT–PCR, total cellular RNA was treated with DNase I and reverse transcribed using oligo(dT) primer. qRT–PCR was carried out using the Maxima SYBR Green/Rox qPCR Master Mix (Fermentas) with gene-specific primers targeting two different exons for forward and reverse primers. PCR was conducted with an Applied Biosystems 7500 apparatus. Relative gene expression for individual genes was normalized to GAPDH.
Preparation and sequencing of RNA-Seq libraries
Total RNA extracted using the RNeasy kit (Qiagen) was first checked for integrity on an Agilent Bioanalyzer 2100; samples with RNA integrity number (RIN) >9.0 were used for subsequent processing. Total RNA was subjected to two rounds of poly(A) selection using oligo-(dT)25 magnetic beads (New England Biolabs) and poly(A)+ RNA was fragmented with NaHCO3 (pH 9.3) for 2 min at 94°C. A single-read (strand specific) cDNA library was prepared following the Illumina TrueSeq small RNA protocol for strand-specific RNA-SeqCitation80 with minor modifications. cDNA was then amplified by PCR for 15 cycles with a universal forward primer and a reverse primer with bar code. The cDNA libraries were sequenced using an Illumina GAIIx instrument. At least 10 million reads per sample were acquired using 70-bp single-end reads.
Bioinformatic and statistical analysis
HTSeqCitation81 was used to convert reads mapped with TopHat in SAM format to gene read counts. Reads were converted to gene counts using the hg19 genome as reference. HTSeq-converted read counts were used as an input for differential gene analysis in the R platform. Read counts were filtered and normalized, and differential gene expression was calculated using edgeR as described in the RUVSeq manual. Sequenced reads were mapped to the human reference genome hg19 using Bowtie 2.Citation82 mRNA expression was measured as RPKM (reads per kilobase of transcript per million reads mapped) at RefSeq transcript level using reads mapped to coding regions, and then summarized at the gene level using the median of measurements by group. Hierarchical clustering was conducted to classify genes and samples based on their expression pattern by Cluster 3.0 and Java TreeView (http://bonsai.hgc.jp/∼mdehoon/software/cluster/software.htm). Differential expression was assessed using both fold change and t-test p-value, which were used to consider both the degree and the significance of regulation.
GO analysis
Biological pathways associated with significant genes were analyzed by Gene Ontology (GO) analysis (http://www.geneontology.org). Mapping between genes and GO entries was obtained from the NCBI Gene database (http://www.ncbi.nlm.nih.gov/gene). The association of genes and pathways was assessed using Fisher's exact test. Redundant concepts in GO results were removed by comparing overlapped portions between genes related to GO entries.
Alternative splicing analysis
Alternative splicing events were detected by the MISO (mixture of isoforms) method.Citation83 Differentially expressed isoforms were further identified according to Bayes factors calculated by MISO and filtering criteria, Psi values (Ψ) and confidence intervals, when comparing the control and knockdown samples. In comparison to the control, all alternative exons presented here changed with Bayes factor ≥ 20.
Disclosure of potential conflicts of interest
The data in part form the basis for patent US 8603814 B2 held by four of the authors: TP, MBM, MH and HMHA.
KTRS_S_1366294.zip
Download Zip (3.7 MB)Acknowledgements
We thank Ms. Anita Antes for technical assistance, and Dr. Leslie Michelson and Dr. Patricia Soteropoulos for discussions and encouragement.
Funding
This work was supported in part by the Foundation of UMDNJ (to MBM), and the National Institutes of Health (HG006339 to MBM and BT, and GM084089 to BT).
References
- Hershey JW, Sonenberg N, Mathews MB. Principles of translational control: an overview. Cold Spring Harb Perspect Biol. 2012;4:1-10. doi:10.1101/cshperspect.a011528. PMID:23209153
- Shoemaker CJ, Green R. Translation drives mRNA quality control. Nat Struct Mol Biol. 2012;19:594-601. doi:10.1038/nsmb.2301. PMID:22664987
- Celik A, Kervestin S, Jacobson A. NMD: At the crossroads between translation termination and ribosome recycling. Biochimie. 2015;114:2-9. doi:10.1016/j.biochi.2014.10.027. PMID:25446649
- Lykke-Andersen S, Jensen TH. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat Rev Mol Cell Biol. 2015;16:665-677. doi:10.1038/nrm4063. PMID:26397022
- Popp MW, Maquat LE. Organizing principles of mammalian nonsense-mediatedmRNA decay. Annu Rev Genet. 2013;47:139-165. doi:10.1146/annurev-genet-111212-133424. PMID:24274751
- Brogna S, McLeod T, Petric M. The meaning of NMD: translate or perish. Trends Genet. 2016;32:395-407. doi:10.1016/j.tig.2016.04.007. PMID:27185236
- Fatscher T, Boehm V, Gehring NH. Mechanism, factors, and physiological role of nonsense-mediated mRNA decay. Cell Mol Life Sci. 2015;72:4523-4544. doi:10.1007/s00018-015-2017-9. PMID:26283621
- Smith JE, Baker KE. Nonsense-mediated RNA decay–a switch and dial for regulating gene expression. Bioessays. 2015;37:612-623. doi:10.1002/bies.201500007. PMID:25820233
- Hamid FM, Makeyev EV. Emerging functions of alternative splicing coupled with nonsense-mediated decay. Biochem Soc Trans. 2014;42:1168-1173. doi:10.1042/BST20140066. PMID:25110020
- Sibley CR. Regulation of gene expression through production of unstable mRNA isoforms. Biochem Soc Trans. 2014;42:1196-1205. doi:10.1042/BST20140102. PMID:25110025
- Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci U S A. 2003;100:189-192. doi:10.1073/pnas.0136770100. PMID:12502788
- Mitrovich QM, Anderson P. Unproductively spliced ribosomal protein mRNAs are natural targets of mRNA surveillance in C. elegans. Genes Dev. 2000;14:2173-2184
- Holbrook JA, Neu-Yilik G, Hentze MW, Kulozik AE. Nonsense-mediated decay approaches the clinic. Nat Genet. 2004;36:801-808. doi:10.1038/ng1403. PMID:15284851
- Lejeune F, Maquat LE. Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells.Curr Opin Cell Biol. 2005;17:309-315. doi:10.1016/j.ceb.2005.03.002. PMID:15901502
- Miller JN, Pearce DA. Nonsense-mediated decay in genetic disease: friend or foe? Mutat Res Rev Mutat Res. 2014;762:52-64. doi:10.1016/j.mrrev.2014.05.001. PMID:25485595
- Frischmeyer PA, Dietz HC. Nonsense-mediated mRNA decay in health and disease. Hum Mol Genet. 1999;8:1893-1900. PMID:10469842
- Mort M, Ivanov D, Cooper DN, Chuzhanova NA. A meta-analysis of nonsense mutations causing human genetic disease. Hum Mutat. 2008;29:1037-1047. doi:10.1002/humu.20763. PMID:18454449
- Keeling KM, Xue X, Gunn G, Bedwell DM. Therapeutics based on stop codon readthrough. Annu Rev Genomics Hum Genet. 2014;15:371-394. doi:10.1146/annurev-genom-091212-153527. PMID:24773318
- Schweingruber C, Rufener SC, Zund D, Yamashita A, Muhlemann O. Nonsense-mediated mRNA decay – mechanisms of substrate mRNA recognition and degradation in mammalian cells. Biochim Biophys Acta. 2013;1829:612-623. doi:10.1016/j.bbagrm.2013.02.005. PMID:23435113
- Hurt JA, Robertson AD, Burge CB. Global analyses of UPF1 binding and function reveal expanded scope of nonsense-mediated mRNA decay. Genome Res. 2013;23:1636-1650. doi:10.1101/gr.157354.113. PMID:23766421
- Nagy E, Maquat LE. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci. 1998;23:198-199. PMID:9644970
- Mathews MB, Hershey JW. The translation factor eIF5A and human cancer. Biochim Biophys Acta. 2015;1849:836-844. doi:10.1016/j.bbagrm.2015.05.002. PMID:25979826
- Dever TE, Gutierrez E, Shin BS. The hypusine-containing translation factor eIF5A. Crit Rev Biochem Mol Biol. 2014;49:413-425. doi:10.3109/10409238.2014.939608. PMID:25029904
- Kemper WM, Berry KW, Merrick WC. Purification and properties of rabbit reticulocyte protein synthesis initiation factors M2Balpha and M2Bbeta. J Biol Chem. 1976;251:5551-5557. PMID:965377
- Benne R, Brown-Luedi ML, Hershey JW. Purification and characterization of protein synthesis initiation factors eIF-1, eIF-4C, eIF-4D, and eIF-5 from rabbit reticulocytes. J Biol Chem. 1978;253:3070-3077. PMID:641055
- Saini P, Eyler DE, Green R, Dever TE. Hypusine-containing protein eIF5A promotes translation elongation. Nature. 2009;459:118-121. doi:10.1038/nature08034. PMID:19424157
- Gutierrez E, Shin BS, Woolstenhulme CJ, Kim JR, Saini P, Buskirk AR, Dever TE, et al. eIF5A promotes translation of polyproline motifs. Mol Cell. 2013;51:35-45. doi:10.1016/j.molcel.2013.04.021. PMID:23727016
- Schuller AP, Wu CC, Dever TE, Buskirk AR, Green R. eIF5A Functions Globally in Translation Elongation and Termination. Mol Cell. 2017;66:194-205 e195. doi:10.1016/j.molcel.2017.03.003. PMID:28392174
- Pelechano V, Alepuz P. eIF5A facilitates translation termination globally and promotes the elongation of many non polyproline-specific tripeptide sequences. Nucleic Acids Research. 2017;45:7326-7338. doi:10.1093/nar/gkx479
- Kang HA, Hershey JW. Effect of initiation factor eIF-5A depletion on protein synthesis and proliferation of Saccharomyces cerevisiae. J Biol Chem. 1994;269:3934-3940. PMID:8307948
- Schrader R, Young C, Kozian D, Hoffmann R, Lottspeich F. Temperature-sensitive eIF5A mutant accumulates transcripts targeted to the nonsense-mediated decay pathway. J Biol Chem. 2006;281:35336-35346. PMID:16987817
- Hanauske-Abel HM, Slowinska B, Zagulska S, Wilson RC, Staiano-Coico L, Hanauske AR, McCaffrey T, Szabo P.. Detection of a sub-set of polysomal mRNAs associated with modulation of hypusine formation at the G1-S boundary. Proposal of a role for eIF-5A in onset of DNA replication. FEBS Lett. 1995;366:92-98. PMID:7789538
- Memin E, Hoque M, Jain MR, Heller DS, Li H, Cracchiolo B, Hanauske-Abel HM, Pe'ery T, Mathews MB. Blocking eIF5A modification in cervical cancer cells alters the expression of cancer-related genes and suppresses cell proliferation. Cancer Res. 2014;74:552-562. doi:10.1158/0008-5472.CAN-13-0474. PMID:24220243
- Fujimura K, Choi S, Wyse M, Strnadel J, Wright T, Klemke R. Eukaryotic translation initiation Factor 5A (EIF5A) regulates pancreatic cancer metastasis by modulating RhoA and Rho-associated Kinase (ROCK) protein expression Levels. J Biol Chem. 2015;290:29907-29919. doi:10.1074/jbc.M115.687418. PMID:26483550
- Mandal A, Mandal S, Park MH. Global quantitative proteomics reveal up-regulation of endoplasmic reticulum stress response proteins upon depletion of eIF5A in HeLa cells. Sci Rep. 2016;6:25795. doi:10.1038/srep25795. PMID:27180817
- Taylor CA, Sun Z, Cliche DO, Ming H, Eshaque B, Jin S, Hopkins MT, Thai B, Thompson JE. Eukaryotic translation initiation factor 5A induces apoptosis in colon cancer cells and associates with the nucleus in response to tumour necrosis factor alpha signalling. Exp Cell Res. 2007;313:437-449. PMID:17187778
- Luchessi AD, Cambiaghi TD, Hirabara SM, Lambertucci RH, Silveira LR, Baptista IL, Moriscot AS, Costa-Neto CM, Curi R. Involvement of eukaryotic translation initiation factor 5A (eIF5A) in skeletal muscle stem cell differentiation. J Cell Physiol. 2009;218:480-489. doi:10.1002/jcp.21619. PMID:19006180
- Hanauske-Abel HM, Park MH, Hanauske AR, Popowicz AM, Lalande M, Folk JE. Inhibition of the G1-S transition of the cell cycle by inhibitors of deoxyhypusine hydroxylation. Biochim Biophys Acta. 1994;1221:115-124. PMID:8148388
- Hofmann W, Reichart B, Ewald A, Müller E, Schmitt I, Stauber RH, Lottspeich F, Jockusch BM, Scheer U, Hauber J, et al. Cofactor requirements for nuclear export of Rev response element (RRE)- and constitutive transport element (CTE)-containing retroviral RNAs. an unexpected role for actin. J Cell Biol. 2001;152:895-910.
- Hoque M, Hanauske-Abel HM, Palumbo P, Saxena D, D'Alliessi Gandolfi D, Park MH, Pe'ery T, Mathews MB. Inhibition of HIV-1 gene expression by Ciclopirox and Deferiprone, drugs that prevent hypusination of eukaryotic initiation factor 5A. Retrovirology. 2009;6:90. doi:10.1186/1742-4690-6-90. PMID:19825182
- Li Y, Fu L, Li JB, Qin Y, Zeng TT, Zhou J, Zeng ZL, Chen J, Cao TT, Ban X, et al. Increased expression of EIF5A2, via hypoxia or gene amplification, contributes to metastasis and angiogenesis of esophageal squamous cell carcinoma. Gastroenterology. 2014;146:1701-1713 e1709. doi:10.1053/j.gastro.2014.02.029. PMID:24561231
- Hanauske-Abel HM, Saxena D, Palumbo PE, Hanauske AR, Luchessi AD, Cambiaghi TD, Hoque M, Spino M, D'Alliessi Gandolfi D, Heller DS, et al. Drug-induced reactivation of apoptosis abrogates HIV-1 infection. PloS One. 2013;8:e74414. doi:10.1371/journal.pone.0074414. PMID:24086341
- Olsen ME, Filone CM, Rozelle D, Mire CE, Agans KN, Hensley L, Connor JH. Polyamines and hypusination are required for ebolavirus gene expression and replication. MBio. 2016;7(4):e00882-16. doi:10.1128/mBio.00882-16
- Bevec D, Jaksche H, Oft M, Wöhl T, Himmelspach M, Pacher A, Schebesta M, Koettnitz K, Dobrovnik M, Csonga R, et al. Inhibition of HIV-1 replication in lymphocytes by mutants of the Rev cofactor eIF-5A. Science. 1996;271:1858-1860. PMID:8596953
- Zuk D, Jacobson A. A single amino acid substitution in yeast eIF-5A results in mRNA stabilization. EMBO J. 1998;17:2914-2925. doi:10.1093/emboj/17.10.2914. PMID:9582285
- Valentini SR, Casolari JM, Oliveira CC, Silver PA, McBride AE. Genetic interactions of yeast eukaryotic translation initiation factor 5A (eIF5A) reveal connections to poly(A)-binding protein and protein kinase C signaling. Genetics. 2002;160:393-405. PMID:11861547
- Rossi D, Galvão FC, Bellato HM, Boldrin PE, Andrews BJ, Valentini SR, Zanelli CF. eIF5A has a function in the cotranslational translocation of proteins into the ER. Amino Acids. 2014;46:645-653. doi:10.1007/s00726-013-1618-6. PMID:24306454
- Zanelli CF, Valentini SR. Pkc1 acts through Zds1 and Gic1 to suppress growth and cell polarity defects of a yeast eIF5A mutant. Genetics. 2005;171:1571-1581. doi:10.1534/genetics.105.048082. PMID:16157662
- Chatterjee I, Gross SR, Kinzy TG, Chen KY. Rapid depletion of mutant eukaryotic initiation factor 5A at restrictive temperature reveals connections to actin cytoskeleton and cell cycle progression. Mol Genet Genomics. 2006;275:264-276. PMID:16408210
- Park MH, Cooper HL, Folk JE. Identification of hypusine, an unusual amino acid, in a protein from human lymphocytes and of spermidine as its biosynthetic precursor. Proc Natl Acad Sci U S A. 1981;78:2869-2873. PMID:6789324
- Cooper HL, Park MH, Folk JE, Safer B, Braverman R. Identification of the hypusine-containing protein hy+ as translation initiation factor eIF-4D. Proc Natl Acad Sci U S A. 1983;80:1854-1857. PMID:6403941
- Park MH. The post-translational synthesis of a polyamine-derived amino acid, hypusine, in the eukaryotic translation initiation factor 5A (eIF5A). J Biochem. 2006;139:161-169. PMID:16452303
- Andrus L, Szabo P, Grady RW, Hanauske AR, Huima-Byron T, Slowinska B, Zagulska S, Hanauske-Abel HM. Antiretroviral effects of deoxyhypusyl hydroxylase inhibitors: a hypusine-dependent host cell mechanism for replication of human immunodeficiency virus type 1 (HIV-1). Biochem Pharmacol. 1998;55:1807-1818. PMID:9714299
- Saxena D, Spino M, Tricta F, Connelly J, Cracchiolo BM, Hanauske AR, D'Alliessi Gandolfi D, Mathews MB, Karn J, Holland B, et al. Drug-Based Lead Discovery: The Novel Ablative Antiretroviral Profile of Deferiprone in HIV-1-infected cells and in HIV-infected treatment-naive subjects of a double-blind, placebo-controlled, randomized exploratory trial. PloS One. 2016;11:e0154842. doi:10.1371/journal.pone.0154842. PMID:27191165
- Groskreutz DJ, Sherf BA, Wood KV, Schenborn ET. Increased expression and convenience with the new pGL3 luciferase reporter vectors. Promega Notes Magazine 1995;50:2-6.
- Sun X, Perlick HA, Dietz HC, Maquat LE. A mutated human homologue to yeast Upf1 protein has a dominant-negative effect on the decay of nonsense-containing mRNAs in mammalian cells. Proc Natl Acad Sci U S A. 1998;95:10009-10014. PMID:9707591
- Woeller CF, Gaspari M, Isken O, Maquat LE. NMD resulting from encephalomyocarditis virus IRES-directed translation initiation seems to be restricted to CBP80/20-bound mRNA. EMBO Rep. 2008;9:446-451. doi:10.1038/embor.2008.36. PMID:18369367
- Duncan RF, Hershey JW. Changes in eIF-4D hypusine modification or abundance are not correlated with translational repression in HeLa cells. J Biol Chem. 1986;261:12903-12906. PMID:3091607
- Wolff EC, Kang KR, Kim YS, Park MH. Posttranslational synthesis of hypusine: evolutionary progression and specificity of the hypusine modification. Amino Acids. 2007;33:341-350. PMID:17476569
- Cuccurese M, Russo G, Russo A, Pietropaolo C. Alternative splicing and nonsense-mediated mRNA decay regulate mammalian ribosomal gene expression. Nucleic Acids Research. 2005;33:5965-5977. doi:10.1093/nar/gki905. PMID:16254077
- Rossbach O, Hung LH, Schreiner S, Grishina I, Heiner M, Hui J, Bindereif A. Auto- and cross-regulation of the hnRNP L proteins by alternative splicing. Mol Cell Biol. 2009;29:1442-1451. doi:10.1128/MCB.01689-08. PMID:19124611
- Hyvonen MT, Uimari A, Keinänen TA, Heikkinen S, Pellinen R, Wahlfors T, Korhonen A, Närvänen A, Wahlfors J, Alhonen L, et al. Polyamine-regulated unproductive splicing and translation of spermidine/spermine N1-acetyltransferase. RNA. 2006;12:1569-1582. doi:10.1261/rna.39806. PMID:16809818
- Hauer C, Sieber J, Schwarzl T, Hollerer I, Curk T, Alleaume AM, Hentze MW, Kulozik A. Exon junction complexes show a distributional bias toward alternatively spliced mRNAs and against mRNAs coding for ribosomal proteins. Cell Rep. 2016;16:1588-1603. doi:10.1016/j.celrep.2016.06.096. PMID:27475226
- Metze S, Herzog VA, Ruepp MD, Muhlemann O. Comparison of EJC-enhanced and EJC-independent NMD in human cells reveals two partially redundant degradation pathways. RNA. 2013;19:1432-1448. doi:10.1261/rna.038893.113. PMID:23962664
- Iadevaia V, Caldarola S, Tino E, Amaldi F, Loreni F. All translation elongation factors and the e, f, and h subunits of translation initiation factor 3 are encoded by 5′-terminal oligopyrimidine (TOP) mRNAs. RNA. 2008;14:1730-1736. doi:10.1261/rna.1037108. PMID:18658124
- Meyuhas O, Kahan T. The race to decipher the top secrets of TOP mRNAs. Biochim Biophys Acta. 2015;1849:801-811. doi:10.1016/j.bbagrm.2014.08.015. PMID:25234618
- Mandal A, Mandal S, Park MH. Genome-wide analyses and functional classification of proline repeat-rich proteins: potential role of eIF5A in eukaryotic evolution. PloS One. 2014;9:e111800. doi:10.1371/journal.pone.0111800. PMID:25364902
- Hershey JW. Expression of initiation factor genes in mammalian cells. Biochimie. 1994;76:847-852. PMID:7880901
- Lipowsky G, Bischoff FR, Schwarzmaier P, Kraft R, Kostka S, Hartmann E, Kutay U, Görlich D. Exportin 4: a mediator of a novel nuclear export pathway in higher eukaryotes. Embo J. 2000;19:4362-4371. PMID:10944119
- Xu A, Jao DL, Chen KY. Identification of mRNA that binds to eukaryotic initiation factor 5A by affinity co-purification and differential display. Biochem J. 2004;384:585-590. PMID:15303967
- Xu A, Chen KY. Hypusine is required for a sequence-specific interaction of eukaryotic initiation factor 5A with postsystematic evolution of ligands by exponential enrichment RNA. J Biol Chem. 2001;276:2555-2561. doi:10.1074/jbc.M008982200. PMID:11060315
- Shi XP, Yin KC, Waxman L. Effects of inhibitors of RNA and protein synthesis on the subcellular distribution of the eukaryotic translation initiation factor, eIF-5A, and the HIV-1 rev protein. Biol Signals. 1997;6:143-149. PMID:9285097
- Melnikov S, Mailliot J, Shin BS, Rigger L, Yusupova G, Micura R, Dever TE, Yusupov M Crystal structure of hypusine-containing translation factor eIF5A bound to a rotated eukaryotic ribosome. Journal of Molecular Biology. 2016;428:3570-3576. doi:10.1016/j.jmb.2016.05.011. PMID:27196944
- Sievert H, Venz S, Platas-Barradas O, Dhople VM, Schaletzky M, Nagel CH, Braig M, Preukschas M, Pällmann N, Bokemeyer C. Protein-protein-interaction Network Organization of the hypusine modification system. Mol Cell Proteomics. 2012;11:1289-1305. doi:10.1074/mcp.M112.019059. PMID:22888148
- Menon B, Gulappa T, Menon KM. Eukaryotic initiation factor 5A plays an essential role in luteinizing hormone receptor regulation. Mol Endocrinol. 2014;28:1796-1806. doi:10.1210/me.2014-1132. PMID:25216047
- Gulappa T, Menon B, Menon KM. Hypusination of eukaryotic initiation factor 5A via cAMP-PKA-ERK1/2 pathway is required for ligand-induced downregulation of LH receptor mRNA expression in the ovary. Mol Cell Endocrinol. 2015;413:90-95. doi:10.1016/j.mce.2015.06.014. PMID:26116232
- Minden MD, Hogge DE, Weir SJ, Kasper J, Webster DA, Patton L, Jitkova Y, Hurren R, Gronda M, Goard CA, et al. Oral ciclopirox olamine displays biological activity in a phase I study in patients with advanced hematologic malignancies. Am J Hematol. 2014;89:363-368. doi:10.1002/ajh.23640. PMID:24273151
- Gonzalez-Hilarion S, Beghyn T, Jia J, Debreuck N, Berte G, Mamchaoui K, Mouly V, Gruenert DC, Déprez B, Lejeune F. Rescue of nonsense mutations by amlexanox in human cells. Orphanet J Rare Dis. 2012;7:58. doi:10.1186/1750-1172-7-58. PMID:22938201
- Homan KT, Wu E, Cannavo A, Koch WJ, Tesmer JJ. Identification and characterization of amlexanox as a G protein-coupled receptor kinase 5 inhibitor. Molecules. 2014;19:16937-16949. doi:10.3390/molecules191016937. PMID:25340299
- Hoque M, Ji Z, Zheng D, Luo W, Li W, You B, Park JY, Yehia G, Tian B. Analysis of alternative cleavage and polyadenylation by 3′ region extraction and deep sequencing. Nat Methods. 2013;10:133-139. doi:10.1038/nmeth.2288. PMID:23241633
- Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166-169. doi:10.1093/bioinformatics/btu638. PMID:25260700
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357-359. doi:10.1038/nmeth.1923. PMID:22388286
- Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods. 2010;7:1009-1015. doi:10.1038/nmeth.1528. PMID:21057496

