Abstract
Objective
This study aimed to assess the impact of colonization status on the outcomes of Acinetobacter spp. bloodstream infection (BSI) and investigate the homology and within-host evolution between colonizing and bloodstream carbapenem-resistant Acinetobacter spp. (CRA) to inform antibiotic therapeutic decisions.
Methods
We analyzed clinical outcomes of 46 hematological patients with Acinetobacter spp. BSI and performed whole-genome sequencing on the remaining CRA isolates.
Results
Among the patients, 39.1% (n=18) had prior Acinetobacter spp. colonization. Colonized patients had higher rates of polymicrobial BSI (50.0% vs 21.4%, P=0.044) and CRA BSI (72.2% vs 17.9%, P<0.001), resulting in elevated inflammatory markers and increased 30-day mortality. Each of the eight pairs of the remaining respiratory colonizing and bloodstream CRA strains belonged to the same genomospecies. Each pair exhibited definitive agreement in at least 21 of the 22 most representative antibiotic susceptibility tests. The minimum spanning tree based on multilocus sequence typing (MLST) and phylogenetic trees based on MLST and single nucleotide polymorphism (SNP) all indicated that each pair shared the same minimum branch. Very few non-synonymous SNPs in genic regions were identified during the transition from respiratory colonization to bloodstream infection, with minimal changes in virulence genes. Homology analysis suggested that CRA BSI originated from colonizing isolates in the respiratory tract.
Conclusion
Strict infection control measures are needed to manage Acinetobacter spp. colonisation in hematological patients. Appropriate empirical therapy can be administered for suspected CRA BSI based on the antimicrobial minimum inhibitory concentration of CRA colonising the respiratory tract.
Introduction
Acinetobacter spp. is a complex genus that has become a common pathogen of nosocomial infections.Citation1,Citation2 Acinetobacter baumannii (A. baumannii) is one of the most significant pathogens causing bloodstream infections (BSIs) associated with Acinetobacter spp. The mortality rate of Acinetobacter spp. BSI has been reported to be between 33–68% within 30 days of diagnosis.Citation3–6 Appropriate treatment decisions and effective preventive measures are necessary to reduce the high mortality rate.
Research has shown that inappropriate empirical antibiotic treatment can lead to adverse outcomes in patients with nosocomial Gram-negative bacilli (GNB), particularly in cases of Acinetobacter spp. infections.Citation3,Citation7–10 Our unpublished data suggested that hematological patients with inappropriate empirical therapy had a mortality rate of up to 66.7%. However, one study indicated that 88% of patients with Acinetobacter spp. BSI initially received inappropriate antibiotic therapy.Citation7 Additionally, colonisation has been proven to be an independent risk factor for bacteremia.Citation11–14 Although several studies have analyzed the risk factors for developing bacteremia in colonised patients,Citation11,Citation15 none has reported the impact of colonisation on the clinical outcomes of subsequent bacteremia. Previous studiesCitation13,Citation16 have compared the homology of colonising and bloodstream isolates, such as Staphylococcus aureus (S. aureus) and Enterobacteriaceae, but few studies have investigated Acinetobacter spp. colonisation. At present, respiratory colonisation provides limited information for clinicians when making appropriate treatment decisions for Acinetobacter spp. BSI.
This study compared clinical outcomes in haematological patients during Acinetobacter spp. BSI stratified by colonisation status. Moreover, we aimed to investigate the homology and within-host evolution between respiratory colonising and bloodstream carbapenem-resistant Acinetobacter spp. (CRA) strains to provide a molecular basis for appropriate empirical antibiotic treatment of bacteremia.
Patients and Methods
Study Design
We included all patients diagnosed with haematological diseases and Acinetobacter spp. BSI between April 2013 and June 2023 at a 766-bed tertiary blood disease hospital in Tianjin, China. We analyzed their clinical outcomes stratified by colonization status. In addition, we compared the homology between colonizing and bloodstream CRA strains to guide appropriate empirical antibiotic treatment.
To achieve this, we first used genome sequencing to identify the genomospecies of the available 21 CRA isolates, which included 8 pairs of respiratory and bloodstream strains from the same infection period. We then compared the antibiotic susceptibility profiles between each pair of CRA strains. Finally, we performed multilocus sequence typing (MLST) and single nucleotide polymorphism (SNP) analyses on each pair of CRA strains to determine genetic relationships and within-host evolution. The Ethics Committee of the Institute of Hematology and Blood Diseases Hospital approved this study. All patients or guardians provided informed written consent per the Declaration of Helsinki.
Definitions
The onset of BSI was defined as the collection date of positive blood culture samples, and laboratory examinations such as procalcitonin (PCT) and C-reactive protein (CRP) were performed within 24 h.Citation17 Patients underwent sputum cultures and were examined for bacterial colonization in the nasal cavity, pharynx, and perianal skin using cotton swabs dipped in 0.9% sterile saline. These procedures were conducted twice a week. Positive blood cultures were assessed as clinically significant or contaminants based on organism type, clinical signs, culture results, and clinical course.Citation18 Bacteremias were considered as polymicrobial when two or more clinically significant microorganisms were isolated in the same set of blood cultures. The definitions for carbapenem-resistant (CR), multidrug-resistant (MDR), and extensively drug-resistant (XDR) Acinetobacter spp. were defined according to previously reported criteria.Citation19 The definitions of sepsis or septic shock followed the 2021 Surviving Sepsis Campaign Guidelines.Citation20 The following cut-off values were used for the primary analysis: 0.5 μg/L for PCT and 10 mg/L for CRP. Previous antibiotic use was defined as the administration of any antibiotic for ≥48 h within the month before the onset of BSI. Empirical antibiotic therapy was defined as any antibiotic administered to febrile patients suspected of having bacteremia before susceptibility results were available. Appropriate empirical antibiotic therapy was defined as the administration of one or more active agents against Acinetobacter spp. at an adequate dose within 24 h after the culture was obtained. Definition of clinical cure: local infection disappeared, body temperature returned to normal, chest CT showed that the shadow of the lung disappeared, and blood indexes (PCT, CRP) returned to normal.
Antimicrobial Susceptibility Testing and Strain Preservation
Antibiotic susceptibility testing was conducted at the hospital’s microbiology laboratory using an automated system, VITEK 2 Compact (Bio m rieux Inc., Hazcwood, Mo, USA). The interpretation of antibiotic susceptibilities followed the guidelines established by the Clinical and Laboratory Standards Institute M100 (YEAR 2023).Citation21 The microbiology laboratory has retained CRA strains from 2017 to the present. This collection comprises 8 pairs of strains, each consisting of both respiratory and bloodstream isolates obtained during the same infection period, along with 5 individual bloodstream CRA strains. The strains were collected using filter paper in a strain storage tube and stored in a refrigerator at −80 °C.
Bacterial Strain and DNA Extraction
Following the revival of the previously preserved CRA strains in the laboratory, a single colony was inoculated onto blood agar medium and incubated at 37°C with 5% CO2 for 20 hours. Genomic DNA was extracted from cell pellets using a Bacteria DNA Kit (OMEGA) following the manufacturer’s instructions. Purified DNA samples underwent quality control assessment, and high-quality DNA samples (OD260/280=1.8~2.0, >6ug) were used for fragment library construction.
Library Construction and Illumina HiSeq Sequencing
Sequencing was conducted by Shanghai Biozeron Biotechnology Co., Ltd. (Shanghai, China). For Illumina paired-end sequencing, a minimum of 1μg of genomic DNA was employed. Paired-end libraries with 400bp insert sizes were prepared following Illumina’s standard protocol. Purified genomic DNA was fragmented, blunt ends generated, adapters ligated, and fragments purified, enriched, and PCR amplified. The qualified Illumina paired-end library was used for Illumina NovaSeq 6000 sequencing (150bp*2).
Genome Assembly
Raw paired-end reads were trimmed and quality controlled using Trimmomatic. Clean data from these quality control processes were used for further analysis. Genome assembly was performed using ABySS with multiple-Kmer parameters, and GapCloser software filled remaining gaps and corrected base polymorphisms. Whole Genome Sequencing (WGS) results facilitated the reclassification of CRA strains in accordance with Genome Taxonomy Database,Citation22 and the project was deposited in GenBank under the accession number PRJNA883531.
MLST Analysis
The housekeeper gene sequences of cpn60, fusA, gltA, pyrG, recA, rplB and rpoB were analysed based on the latest pubmlst database, according to the Pasteur multilocus sequence typing (MLST) scheme, to obtain the allelic profiles and sequence types (STs). The phylogenetic tree based on MLST was constructed by RAxML-NGv.0.9.0, and the clusters and minimum spanning tree based on MLST were constructed by BioNumerics.
SNP Analysis
We aligned each sample to the reference sequence using MUMmer software and identified potential single nucleotide polymorphism (SNP) sites. To verify and filter the SNP sites, we compared the extracted sequence with the assembly results using BLAT software. We obtained reliable SNPs and constructed phylogenetic trees by the maximum likelihood method using PhyML software.Citation23 Coding proteins were functionally classified according to Clusters of Orthologous Groups of proteins (COG) database.
Statistical Analysis
Statistical Product and Service Solutions (SPSS) software (version 24.0; Chicago, IL, USA) was used to analyse the data. Categorical variables were compared using the chi-squared or Fisher’s exact tests. Continuous variables were expressed as the median and interquartile range (IQR), and differences were identified using the two-sample t-test or Mann–Whitney U-test. The Kaplan-Meier method was used to plot survival curves (Log rank test). Statistical significance was set at P-values <0.05.
Results
Clinical Outcomes Stratified by Colonization Status
This study retrospectively analysed 46 patients diagnosed with haematological diseases and Acinetobacter spp. BSI between 2013 and 2023. Of 46 patients, 50.0% were male, with a median age of 25.5 years (range: 1–62 years) (Table S1). The underlying diseases included very severe aplastic anaemia, acute lymphoblastic leukaemia, acute myeloid leukaemia, myelodysplastic syndrome, diffuse large B-cell lymphoma and β-thalassemia. summarizes the clinical outcomes of hematological patients with Acinetobacter spp. BSI based on their colonization status. Of the total patients, 39.1% (n=18) had prior Acinetobacter spp. colonisation, with one in the perianal area and the remaining in the respiratory tract. These colonized patients had significantly higher median levels of PCT (median, 1.46 [IQR, 0.71–8.32] vs median, 0.17 [IQR, 0.09–0.69]; P=0.008) and CRP (median, 92.29 [IQR, 16.50–194.00] vs median, 12.88 [IQR, 5.65–51.85]; P=0.013) during their Acinetobacter spp. BSI. Furthermore, they were at an increased risk of developing polymicrobial BSI (50.0% vs 21.4%, P=0.044). During their bacteremia, they also had higher rates of respiratory failure (38.9% vs 10.7%, P=0.033) and sepsis or septic shock (44.4% vs 10.7%, P=0.014).
Table 1 Comparison of Outcomes in Hematological Patients During Acinetobacter Spp. Bloodstream Infection, Stratified by Acinetobacter Spp. Colonization Status
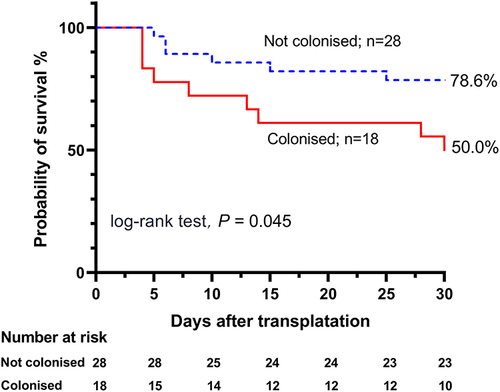
The present study also found that Acinetobacter spp. colonization increased the risk of subsequent MDR Acinetobacter spp. (MDRA) BSI (77.8% vs 25.0%, P<0.001). Among patients with colonization, the bloodstream isolates had significantly higher rates of drug resistance to carbapenems, fluoroquinolones, aminoglycosides and piperacillin-tazobactam (72.2%, 50.0%, 50.0% and 77.8%, respectively) than those without colonization. XDR Acinetobacter spp. (XDRA) BSI (22.2% vs 3.6%, P=0.069) was marginally common in the colonized patients. Additionally, colonized patients were more likely to receive inappropriate empirical antibiotic therapy within 24 hours (66.7% vs 14.3%, P<0.001). Notably, colonized patients had a significantly lower 30-day clinical cure rate (22.2% vs 64.3%, P=0.005) and higher 30-day mortality rate (50.0% vs 21.4%, P=0.044) after the onset of bacteremia. The survival analysis revealed a significant difference in the 30-day survival probability between patients with previous Acinetobacter spp. colonization and those with a negative colonization status (78.6% [95% CI: 67.4–98.7%] vs 50.0% [95% CI: 30.0–74.1%], P=0.045) ().
Antibiotic Susceptibility Profiles and Genomospecies
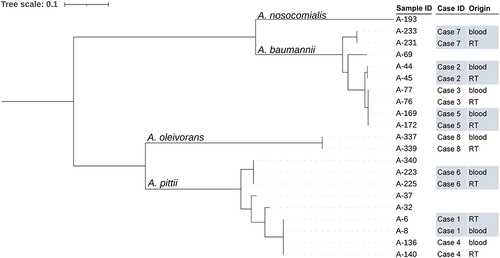
The majority of CRA isolates in our study were identified as A. baumannii or the A. calcoaceticus-baumannii (Acb) complex using VITEK 2 Compact. We observed almost identical antibiotic susceptibility profiles for each of the eight pairs of respiratory-bloodstream CRA strains (Table S2). These profiles showed definitive agreement for at least 21 of the 22 most representative antibiotic susceptibility tests. Therefore, we resuscitated a total of 21 CRA strains that were accessible in the microbiology laboratory. This collection comprised 8 pairs of strains, each consisting of respiratory and bloodstream isolates obtained during the same infection period, as well as 5 individual bloodstream isolates. The genome sequencing revealed the identification of A. pittii (nine), A. baumannii (nine), A. oleivorans (two), and A. nosocomialis (one). We also determined that each of the eight pairs of respiratory and bloodstream CRA strains from the same infection period belonged to the same genomospecies ().
Table 2 The Sequence Types of 21 Carbapenem-Resistant Acinetobacter Spp. Isolates and Four Reference Types Using the Pasteur Multilocus Sequence Typing (MLST) Scheme
High Genetic Similarity Based on MLST and SNP
displays the sequence types (STs) of the CRA strains, as determined by the Pasteur MLST scheme. In addition to the seven known STs, we identified five new housekeeping genes and four new STs that have not been reported internationally. To determine genetic similarity, we conducted phylogenetic analyses based on genome sequencing. The MLST phylogenetic tree demonstrated that each of the eight pairs of respiratory-bloodstream CRA strains shared the same minimum branch (). The MLST minimum spanning tree () connected isolates within the same clonal complexes with solid black lines, indicating their closest kinship. The respiratory-bloodstream CRA strains of case 6 were grouped into a clonal complex, whereas each of the other seven pairs of respiratory-bloodstream CRA strains exhibited an identical MLST pattern.
Figure 2 The phylogenetic tree of 21 carbapenem-resistant Acinetobacter spp. strains and 4 type strains inferred from a concatenate of the seven alleles used in the Pasteur multilocus sequence typing (MLST) scheme. RT, respiratory tract.
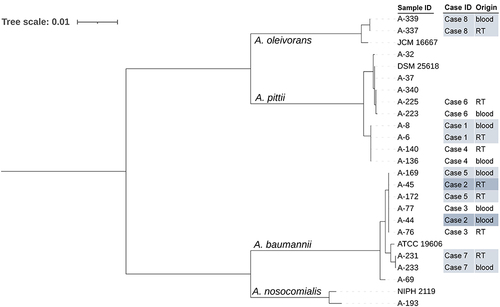
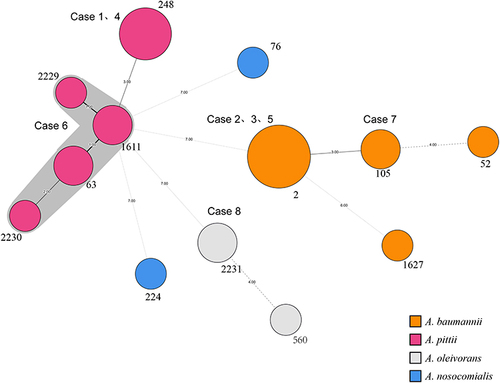
Figure 3 The minimum spanning tree of 21 carbapenem-resistant Acinetobacter spp. (CRA) strains and 4 type strains inferred from a concatenate of the seven alleles used in the Pasteur multilocus sequence typing (MLST) scheme. The sequence types (STs) are indicated by the numbers beside each circle, with the size of each circle proportional to the number of isolates belonging to the same ST type. Branch values indicate the number of loci that differ between adjacent nodes. Grey shading is used to represent the same cluster. Each of the eight pairs of respiratory-bloodstream CRA strains had an identical MLST pattern or was grouped into a clonal complex.
Considering that ST-2 is a prevalent epidemic strain worldwide,Citation24,Citation25 we conducted a comparative genomics analysis based on the complete genome sequence of strain A-169 (A. baumannii, ST-2). A total of 348,540 SNPs were identified in all bloodstream CRA isolates by comparing them with the genome of strain A-169. Consistent with the MLST findings, each pair of respiratory-bloodstream CRA strains from the same patient was classified into one clade in the SNP-based phylogenetic tree, indicating their high degree of genetic similarity (). Collectively, the homology analysis results above suggest that these respiratory strains were likely the direct origin of CRA BSI.
Limited Within-Host Evolution
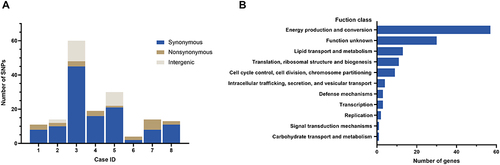
To investigate the genomic evolution from respiratory tract carriage to bloodstream infection, we analyzed SNPs between colonising and bloodstream CRA strains from eight patients with CRA BSI. On average, each bloodstream isolate carried 21 SNPs in 3 to 7 genes, and synonymous SNPs occurred more frequently than non-synonymous SNPs in most patients with a ratio of 5.5:1 (). Functional analysis revealed that most mutations were enriched in genes associated with energy production and conversion, lipid transport and metabolism, and translation, ribosomal structure and biogenesis. ().
Figure 5 (A) The genic and intergenic single nucleotide polymorphisms (SNPs) identified during within-host evolution. (B) Function class of genes with all SNPs based on the Clusters of Orthologous Groups of proteins (COG).
We identified all non-synonymous genic SNPs and determined the corresponding base and amino acid changes. summarizes the names or functions and the function classes of the corresponding genes. We found a nonsense mutation on rnd, which encodes ribonuclease D, and a stop codon mutation on pdaA, which encodes polysaccharide deacetylase. SNPs were also found in ahpF, TMP, pdaA and Ata genes, which encode alkyl hydroperoxide reductase subunit F, phage tail tape measure protein, polysaccharide deacetylase, and autotransporter adhesin, respectively. Notably, a mutation in the housekeeper gene rpo was detected in one patient (case 6) during the breakthrough into the bloodstream, resulting in a change in sequence type from ST1611 to ST2229. Only one non-synonymous SNP was detected in the virulence genes (Ata gene coding autotransporter adhesin).
Table 3 Non-Synonymous Genic Single Nucleotide Polymorphisms (SNPs) Found from Respiratory Tract Carriage to Bloodstream Infection in Eight Patients with Carbapenem-Resistant Acinetobacter Spp.
Discussion
We observe that the prevalence of Acinetobacter spp. BSI among patients with hematologic disorders is low and has been infrequently documented. A multicenter study conducted from January 2014 to June 2015 in the hematology wards of 18 tertiary hospitals in China reported that A. baumannii bacteremia accounted for only 2.9% (40/1358) of all cases of bacteremia.Citation26 In our own study, we included a total of 46 patients with Acinetobacter spp. BSI over a 10-year follow-up period in a large hematology hospital. Nevertheless, it is essential to emphasize the high mortality rate associated with hematologic patients who have concurrent Acinetobacter spp. BSI, warranting careful consideration.
In the last few years, A. pittii, A. calcoaccius, A. lwoffii, A. junii, A. soli, A. ursingii, A. bereziniae and A. nosocomialis have gradually emerged as common pathogens of nosocomial infection.Citation1,Citation2 A. oleivorans, traditionally recognized as an oil-degrading bacterium and a focus of environmental engineering studies, has recently been identified as a human pathogen as well.Citation27,Citation28 The propensity of Acinetobacter spp. to colonize and infect critically ill patients often leads to a poor prognosis.Citation7 It has been widely reported that selective pressure from colonisation is an independent risk factor associated with breakthrough Gram-negative bacteremia,Citation11–14,Citation29 particularly during carbapenem therapy.Citation12 Empirical antibiotic therapy is a critical component in the treatment of Acinetobacter spp. BSI.Citation26 In a large multicenter study, inappropriate empirical antibiotic therapy was found to nearly doubled hospital mortality in patients with A. baumannii pneumonia and sepsis (adjusted RRR 1.8, 95% CI 1.4–2.3, P<0.001).Citation8 Therefore, optimizing empirical antibiotic therapy for patients with Acinetobacter spp. BSI is a pressing concern, given the increasing resistance rates to existing antibiotics.
According to a study,Citation30 polymicrobial A. baumannii BSI accounted for 19.1% (39/204) of all A. baumannii BSI cases. Moreover, the resistance rate of A. baumannii to imipenem, cefepime, tobramycin, piperacillin/tazobactam, and ciprofloxacin was significantly higher in the polymicrobial A. baumannii BSI group than in the monomicrobial A. baumannii BSI group. While colonization pressure has been suggested as a promoter of acquiring MDR bacteria, it has not been widely studied for Acinetobacter spp.Citation31,Citation32 In the present study, we found that the colonized patients had an increased risk of developing polymicrobial BSI. Additionally, Acinetobacter spp. colonization significantly increased the resistance rates of Acinetobacter spp. BSI to multiple antibiotics. Consequently, the increased incidence of CRA and MDRA BSI in our study raised the possibility of patients receiving inappropriate empirical antibiotic therapy, leading to higher levels of inflammatory indicators and higher 30-day mortality. Based on our findings, colistin appears to be the most effective therapeutic option against the isolates identified in our study.
There have been limited investigations into the molecular correlation between colonizing and bloodstream Acinetobacter spp. Moreover, clinical bacterial identification methods often classify Acinetobacter spp. strains as Acb complex or A. baumannii due to their limitations.Citation33 Popular molecular epidemiology methods, such as MLST, pulsed-field gel electrophoresis (PFGE) and WGS, can be used to identify genetic similarity.Citation34 For instance, Johanna et alCitation13 performed PFGE on nine hemodialysis patients colonized by S. aureus who later presented with bacteremia caused by the same bacteria. The Dice index revealed that 77.8% of patients were infected with the same strain previously identified as colonizing, with 100% similarity. Similarly, Michael et alCitation16 confirmed that each of the ten pairs of colonizing and bloodstream isolates from patients with extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae bacteremia had identical MLST patterns and ESBL genes. Additionally, PFGE indicated a genetic similarity of >95%. In our study, we found that each of the eight pairs of colonizing and bloodstream CRA strains had identical genomospecies using gene sequence, with nearly identical antimicrobial susceptibility profiles for 22 tested antibiotics. Furthermore, in the MLST-based phylogenetic tree, MLST-based minimum spanning tree, and the SNP-based phylogenetic tree, each pair was divided into the same minimum branch, suggesting close genetic relatedness.
To gain a better understanding of the molecular mechanisms underlying within-host evolution of CRA BSI, we identified SNPs that accompanied the transition from respiratory tract colonization to bloodstream infection. In all cases, colonization progressed to bacteremia, with only one non-synonymous SNP detected in virulence genes. Specifically, the gene Ata, which plays a pivotal role in host adherence by recognizing host glycans as high-affinity receptors was found to be affected.Citation35 Another studyCitation36 that aimed to identify genomic modifications occurring in S. aureus isolates colonizing the nares as they progressed to bacteremia in eight patients did not reveal the addition of new virulence genes. Our findings suggested that the CRA genome remained relatively stable during the transition from respiratory tract colonization to bloodstream infection, and did not undergo frequent genetic recombination or clonal selection, which was consistent with a genomic analysis of consecutive A. baumannii strains from a single patient.Citation37
To the best of our knowledge, this is the first study to examine the homology between colonizing and bloodstream CRA strains. However, the retrospective nature of the study limited our ability to comprehensively assess patients, as inclusion relied on physician clinical judgment. Additionally, the stress of freezing may have altered microbiological features.
In conclusion, our analysis of clinical data suggested that Acinetobacter spp. colonization increased the risk of subsequent antibiotic-resistant Acinetobacter spp. BSI, leading to elevated inflammation markers during bacteremia and higher 30-day mortality. Strict infection control measures are necessary to manage Acinetobacter spp. colonisation in haematological patients. Moreover, the identical genomospecies, nearly identical drug-resistance profiles, high genetic similarity based on MLST and SNP analysis, and limited within-host evolution indicated that these patients developed CRA BSI from colonizing CRA isolates in the respiratory tract. Therefore, appropriate empirical therapy can be administered for suspected CRA BSI based on the antimicrobial minimum inhibitory concentration of CRA colonising the respiratory tract.
Patient Consent Statement
We have obtained written informed consent from the patient or patient’s parent/guardian.
Data Sharing Statement
The datasets generated and/or analysed during the current study are available in the GenBank repository. You can access them through the following web link: https://www.ncbi.nlm.nih.gov/nuccore/?term=PRJNA883531, and the accession number is PRJNA883531.
Ethics Approval Statement
The study design was approved by the Ethics Committee of the Blood Diseases Hospital, Chinese Academy of Medical Sciences. Lot number: IIT2022071-EC-1. Please refer to the attached file.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
Acknowledgments
Thanks are due to Fupin Hu and Siquan Shen for bioinformatics analysis.
Additional information
Funding
References
- Tavares LCB, Cunha MPV, De Vasconcellos FM, et al. Genomic and clinical characterization of IMP-1-producing multidrug-resistant Acinetobacter bereziniae isolates from bloodstream infections in a Brazilian tertiary hospital. Microb Drug Resist. 2020;26(11):1399–1404. doi:10.1089/mdr.2019.0210
- Baraka A, Traglia GM, Montana S, et al. An Acinetobacter non-baumannii population study: antimicrobial resistance genes (ARGs). Antibiotics. 2020;10(1). doi:10.3390/antibiotics10010016
- Lee HY, Chen CL, Wu SR, et al. Risk factors and outcome analysis of Acinetobacter baumannii complex bacteremia in critical patients. Crit Care Med. 2014;42(5):1081–1088. doi:10.1097/CCM.0000000000000125
- Wisplinghoff H, Paulus T, Lugenheim M, et al. Nosocomial bloodstream infections due to Acinetobacter baumannii, Acinetobacter pittii and Acinetobacter nosocomialis in the United States. J Infect. 2012;64(3):282–290. doi:10.1016/j.jinf.2011.12.008
- Balkhair A, Al-Muharrmi Z, Al’adawi B, et al. Prevalence and 30-day all-cause mortality of carbapenem-and colistin-resistant bacteraemia caused by Acinetobacter baumannii, pseudomonas aeruginosa, and Klebsiella pneumoniae: description of a decade-long trend. Int J Infect Dis. 2019;85:10–15. doi:10.1016/j.ijid.2019.05.004
- Freire MP, De Oliveira Garcia D, Garcia CP, et al. Bloodstream infection caused by extensively drug-resistant Acinetobacter baumannii in cancer patients: high mortality associated with delayed treatment rather than with the degree of neutropenia. Clin Microbiol Infect. 2016;22(4):352–358. doi:10.1016/j.cmi.2015.12.010
- Leao AC, Menezes PR, Oliveira MS, et al. Acinetobacter spp. are associated with a higher mortality in intensive care patients with bacteremia: a survival analysis. BMC Infect Dis. 2016;16:386. doi:10.1186/s12879-016-1695-8
- Zilberberg MD, Nathanson BH, Sulham K, et al. Multidrug resistance, inappropriate empiric therapy, and hospital mortality in Acinetobacter baumannii pneumonia and sepsis. Crit Care. 2016;20(1):221. doi:10.1186/s13054-016-1392-4
- Zasowski EJ, Bassetti M, Blasi F, et al. A systematic review of the effect of delayed appropriate antibiotic treatment on the outcomes of patients with severe bacterial infections. Chest. 2020;158(3):929–938. doi:10.1016/j.chest.2020.03.087
- Choi SH, Cho EB, Chung JW, et al. Changes in the early mortality of adult patients with carbapenem-resistant Acinetobacter baumannii bacteremia during 11 years at an academic medical center. J Infect Chemother. 2019;25(1):6–11. doi:10.1016/j.jiac.2018.09.011
- Kim SY, Cho SI, Bang JH. Risk factors associated with bloodstream infection among patients colonized by multidrug-resistant Acinetobacter baumannii: a 7-year observational study in a general hospital. Am J Infect Control. 2020;48(5):581–583. doi:10.1016/j.ajic.2019.07.025
- Lee JY, Kang CI, Ko JH, et al. Clinical features and risk factors for development of breakthrough gram-negative bacteremia during carbapenem therapy. Antimicrob Agents Chemother. 2016;60(11):6673–6678. doi:10.1128/AAC.00984-16
- Vanegas JM, Salazar-Ospina L, Roncancio GE, et al. Staphylococcus aureus colonization increases the risk of bacteremia in hemodialysis patients: a molecular epidemiology approach with time-dependent analysis. Am J Infect Control. 2021;49(2):215–223. doi:10.1016/j.ajic.2020.05.031
- Silago V, Kovacs D, Msanga DR, et al. Bacteremia in critical care units at Bugando Medical Centre, Mwanza, Tanzania: the role of colonization and contaminated cots and mothers’ hands in cross-transmission of multidrug resistant Gram-negative bacteria. Antimicrob Resist Infect Control. 2020;9(1):58. doi:10.1186/s13756-020-00721-w
- Jung JY, Park MS, Kim SE, et al. Risk factors for multi-drug resistant Acinetobacter baumannii bacteremia in patients with colonization in the intensive care unit. BMC Infect Dis. 2010;10:228. doi:10.1186/1471-2334-10-228
- Satlin MJ, Chavda KD, Baker TM, et al. Colonization with levofloxacin-resistant extended-spectrum beta-lactamase-producing Enterobacteriaceae and risk of bacteremia in hematopoietic stem cell transplant recipients. Clin Infect Dis. 2018;67(11):1720–1728. doi:10.1093/cid/ciy363
- Chiang DH, Wang CC, Kuo HY, et al. Risk factors for mortality in patients with Acinetobacter baumannii bloodstream infection with genotypic species identification. J Microbiol Immunol Infect. 2008;41(5):397–402.
- Diekema DJ, Beekmann SE, Chapin KC, et al. Epidemiology and outcome of nosocomial and community-onset bloodstream infection. J Clin Microbiol. 2003;41(8):3655–3660. doi:10.1128/JCM.41.8.3655-3660.2003
- Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi:10.1111/j.1469-0691.2011.03570.x
- Evans L, Rhodes A, Alhazzani W, et al. Surviving sepsis campaign: international guidelines for management of sepsis and septic shock 2021. Crit Care Med. 2021;49(11):e1063–e1143. doi:10.1097/CCM.0000000000005337
- Pierce VM, Bhowmick T, Simner PJ. Guiding antimicrobial stewardship through thoughtful antimicrobial susceptibility testing and reporting strategies: an updated approach in 2023. J Clin Microbiol. 2023;61(11):e0007422. doi:10.1128/jcm.00074-22
- Rinke C, Chuvochina M, Mussig AJ, et al. A standardized archaeal taxonomy for the genome taxonomy database. Nat Microbiol. 2021;6(7):946–959. doi:10.1038/s41564-021-00918-8
- Tan KK, Tan YC, Chang LY, et al. Full genome SNP-based phylogenetic analysis reveals the origin and global spread of Brucella melitensis. BMC Genomics. 2015;16(1):93. doi:10.1186/s12864-015-1294-x
- Tomaschek F, Higgins PG, Stefanik D, et al. Head-to-head comparison of two multi-locus sequence typing (MLST) schemes for characterization of Acinetobacter baumannii outbreak and sporadic isolates. PLoS One. 2016;11(4):e0153014. doi:10.1371/journal.pone.0153014
- Khurshid M, Rasool MH, Ashfaq UA, et al. Dissemination of blaOXA-23-harbouring carbapenem-resistant Acinetobacter baumannii clones in Pakistan. J Glob Antimicrob Resist. 2020;21:357–362. doi:10.1016/j.jgar.2020.01.001
- Wang X, Zhang L, Sun A, et al. Acinetobacter baumannii bacteraemia in patients with haematological malignancy: a multicentre retrospective study from the infection working party of Jiangsu society of hematology. Eur J Clin Microbiol Infect Dis. 2017;36(7):1073–1081. doi:10.1007/s10096-016-2895-2
- Li J, Feng X, Wang J, et al. Acinetobacter spp. bloodstream infection in hematological patients: a 10-year single-center study. BMC Infect Dis. 2023;23(1):796. doi:10.1186/s12879-023-08789-6
- Sheck E, Romanov A, Shapovalova V, et al. Acinetobacter non-baumannii species: occurrence in infections in hospitalized patients, identification, and antibiotic resistance. Antibiotics. 2023;12(8):1.
- Jang TN, Lee SH, Huang CH, et al. Risk factors and impact of nosocomial Acinetobacter baumannii bloodstream infections in the adult intensive care unit: a case-control study. J Hosp Infect. 2009;73(2):143–150. doi:10.1016/j.jhin.2009.06.007
- Chen Q, Zheng Z, Shi Q, et al. Multidrug-resistant Acinetobacter baumannii may cause patients to develop polymicrobial bloodstream infection. Can J Infect Dis Med Microbiol. 2022;2022:8368578. doi:10.1155/2022/8368578
- Playford EG, Craig JC, Iredell JR. Carbapenem-resistant Acinetobacter baumannii in intensive care unit patients: risk factors for acquisition, infection and their consequences. J Hosp Infect. 2007;65(3):204–211. doi:10.1016/j.jhin.2006.11.010
- Arvaniti K, Lathyris D, Ruimy R, et al. The importance of colonization pressure in multiresistant Acinetobacter baumannii acquisition in a Greek intensive care unit. Crit Care. 2012;16(3):R102. doi:10.1186/cc11383
- E TB, Paterson DL, Kamolvit W, et al. Species identification within Acinetobacter calcoaceticus-baumannii complex using MALDI-TOF MS. J Microbiol Methods. 2015;118:128–132. doi:10.1016/j.mimet.2015.09.006
- Mao P, Deng X, Yan L, et al. Whole-genome sequencing elucidates the epidemiology of multidrug-resistant Acinetobacter baumannii in an intensive care unit. Front Microbiol. 2021;12:715568. doi:10.3389/fmicb.2021.715568
- Tram G, Poole J, Adams FG, et al. The Acinetobacter baumannii autotransporter adhesin ata recognizes host glycans as high-affinity receptors. ACS Infect Dis. 2021;7(8):2352–2361. doi:10.1021/acsinfecdis.1c00021
- Benoit JB, Frank DN, Bessesen MT. Genomic evolution of Staphylococcus aureus isolates colonizing the nares and progressing to bacteremia. PLoS One. 2018;13(5):e0195860. doi:10.1371/journal.pone.0195860
- Kim SJ, Kim YJ, Ko KS. Genomic analysis of consecutive Acinetobacter baumannii strains from a single patient. Front Microbiol. 2018;9:2840. doi:10.3389/fmicb.2018.02840