Abstract
Background. Leucine-rich and immunoglobulin-like domains 1-3 (LRIG1-3) proteins have been implicated in the regulation of EGFR signalling. In the present study, we investigated the clinical implications of the expression of EGFR and LRIG1-3 in oesophageal carcinoma, as well as the correlation between their expression levels and the chemosensitivity of oesophageal carcinoma cell lines. Patients and methods. Tumours from 80 patients with oesophageal carcinoma were investigated for the expression of EGFR and LRIG proteins by immunohistochemistry. Oesophageal carcinoma cell lines were investigated for their expression of EGFR and LRIG1, 2, and 3 by quantitative real time RT-PCR and for their sensitivity to commonly used chemotherapeutics by a cytotoxicity assay. Results and discussion: Based on a total score of intensity and expression rates, a trend towards survival difference was found for EGFR (p = 0.09) and LRIG2 (p = 0.18) whereas for LRIG1 and -3 there was no trend towards any association with survival. Correlation analysis revealed a correlation with the clinical expression of EGFR and LRIG3 (p = 0.0007). Significant correlations were found between LRIG1 expression levels and sensitivity to cisplatin (r = −0.74), docetaxel (r = −0.69), and vinorelbine (r = −0.82) in oesophageal carcinoma cell lines. EGFR and the LRIG proteins may be functionally involved in oesophageal carcinoma, but larger materials are needed to fully elucidate the clinical implication.
Oesophageal carcinoma is the seventh most common cause of cancer-related death in the western world and the incidence is increasing [Citation1]. The major challenge in the treatment of oesophageal carcinoma is to reduce the risk of local recurrence. Following surgical resection, local tumour recurrence occurs in approximately 50% of all patients. Most of these recurrences occur within the first year after surgery, and the prognosis for these patients is dismal [Citation2]. For patients treated with curatively intended radiotherapy or radiochemotherapy (including radiation doses at the limit of normal-tissue tolerance), approximately 80% relapse at the primary site [Citation3]. Despite new treatment modalities, survival remains poor for patients with oesophageal carcinoma, and treatment-related toxicity can be devastating. Consequently, selecting appropriate patients for the different treatment modalities is crucial, as is identifying predictive factors for response to therapy.
The epidermal growth factor (EGF) receptor (EGFR) family members, EGFR (also called ERBB1), ERBB2 (also called HER2 or Neu), ERBB3, and ERBB4 and their ligands are important for the development and progression of various malignancies [Citation4]. In oesophageal carcinoma, EGFR has been reported to be commonly overexpressed [Citation5], and the overexpression seems to be independent of mutations in TP53, RAS, or other investigated oncogenes. The signal cascade system of EGFR is involved in oesophageal tumour growth, and EGFR-targeted therapies have been shown to be capable of suppressing this growth [Citation6].
Leucine-rich and immunoglobulin-like domains (LRIG) 1, 2, and 3 are integral membrane proteins containing an extracellular or luminal region consisting of a leucine-rich repeats domain and three immunoglobulin-like domains, a transmembrane domain, and a cytosolic tail [Citation7–10]. LRIG1 has been postulated to be a tumour suppressor [Citation11] and shown to counteract the signalling of EGFR [Citation12,Citation13], MET [Citation14], and RET [Citation15] receptors. LRIG1 participates in an EGF-driven negative feedback loop involving receptor ubiquitination and degradation leading to the suppression of EGFR receptor signalling. Less is known about the function of the other two human LRIG paralogs, LRIG2 and LRIG3. The location of the LRIG1, 2, and 3 genes at chromosomes 3p14, 1p13, and 12q13, respectively, also indicates interesting functions in different tumour forms [Citation7–9]. LRIG1 is expressed in most or all tissues [Citation8] and has also been suggested to be of prognostic significance in several malignancies [Citation11]. For example, LRIG1 expression is down-regulated in renal cell carcinoma [Citation16], and high LRIG1 expression is associated with long survival in squamous cell carcinoma of the skin and cervix [Citation17,Citation18]. LRIG2 expression, on the other hand, has been associated with short survival in oligodendroglioma [Citation19] and squamous cell carcinoma of the uterine cervix [Citation20]. In the present study we investigated the clinical implications of EGFR and LRIG1-3 expression in oesophageal carcinoma with the aim of investigating their clinical prognostic effect. Furthermore, the importance of the expression levels of the EGFR and LRIG family members for the sensitivity of oesophageal carcinoma cell lines for standard chemotherapeutic agents was analysed.
Patients and methods
Patients
Between 1990 and 2000, 126 patients were recorded as having received treatment for oesophageal carcinoma at the Department of Oncology, Uppsala University Hospital, Sweden. Treatment strategies for these patients were preoperative chemotherapy and radiation treatment followed by surgery, curatively intended radiation treatment or palliative treatment including radiation treatment or chemotherapy. From 80 of these patients, formalin-fixed and paraffin- embedded tumour samples were obtained for immunohistochemical analysis of EGFR and LRIG1-3. The following clinical parameters were evaluated in the 80 patients: age; gender; performance status at first admittance; smoking history; tumour localisation grouped into upper (15–24 cm), middle (25–34 cm), and lower (35–46 cm) part of oesophagus; tumour histology; and tumour stage at first admittance, defined as localised or metastatic disease. No data were available concerning surgical resection grade or surgically related complications. Collection of the clinical parameters started when the patient was first admitted to an Oncology Department. The patients were followed until March 12, 2003 and mean survival in the material was 547 days (median = 266 days, min = 5 days, max = 3675 days). The study was reviewed and approved by the research ethics committee, Uppsala University, Uppsala, Sweden.
Immunohistochemistry
The immunohistochemical staining procedures have previously been described for EGFR [Citation21], LRIG1 [Citation8], LRIG2 [Citation22], and LRIG3 [Citation23]. Briefly, 4 μm-thick tissue sections were labelled with the following primary antibodies at the indicated concentrations: anti-EGFR 1005 (2 μg/ml, Santa Cruz Biotechnology, Santa Cruz, CA, USA,), anti-LRIG1 (0.5 μg/ml, AgriSera, Vännäs, Sweden), anti-LRIG2 (1 μg/ml), and anti-LRIG3 (2.2 μg/ml). Immunodetection was achieved by using horseradish peroxidase-conjugated secondary antibodies and the substrate diaminobenzidine. The slides were counterstained with haematoxylin and mounted in glycerol-gelatin.
Scoring of immunostainings
The evaluation of the immunostaining was blinded with respect to the clinical parameters of the patients and performed by two independent observers. Immunostainings were scored as previously described [Citation24]. In brief, positive immunohistochemical staining in tumour cells was evaluated with respect to the fraction of positive tumour cells and the intensity of positive staining. From these scoring data, three categories were defined: Grade 3: Strong immunoreactivity in >25% of tumour cells; Grade 2: moderate immunoreactivity in >25% of tumour cells, or strong immunoreactivity in <25% of tumour cells; Grade 1: weak immunoreactivity in >25% of tumour cells, or moderate immunoreactivity in <25% of tumour cells; Grade 0: Lack of immunostaining (negative), or weak immunostaining in <25% of tumour cells. A total score based on low fraction and intensity (graded as 0 or 1) versus high fraction and intensity (graded 2 or 3) was created for each patient and statistically analysed.
The subcellular localisation of the stainings was evaluated and found to be predominantly cyoplasmic in all stainings. The different tumour stainings were compared to each other and used as internal controls.
Cell culturing
The human oesophageal squamous cell carcinoma cell lines KYSE30, KYSE70, KYSE140, KYSE150, KYSE180, KYSE410, KYSE450, KYSE510, and KYSE520 (Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany) were cultivated in RPMI 1640 medium, supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 50 U/ml penicillin, and 100 μg/ml streptomycin (Sigma-Aldrich Sweden AB, Stockholm, Sweden) in 37°C humidified air with 5% CO2. Cells were sub-cultivated twice weekly and harvested in log-phase for experimental use.
Cytotoxicity assay
The fluorometric microculture cytotoxicity assay (FMCA) [Citation25] was used to investigate the in vitro effect of seven clinically used cytotoxic drugs: 5fluorouracil (5FU), cisplatin, docetaxel, gemcitabine, etoposide, melphalan, and vinorelbine (all from Swedish National Pharmacy). Briefly, 384-well microtiter plates (Nunclon surface, NUNC Brand Products, Roskilde, Denmark) were prepared with duplicates of 5 μl drug solutions at five different concentrations (five-fold serial dilutions). Cells (1 × 105 cells/ml; 45 μl) were seeded into the drug-prepared microtiter plates at a cell density of 1 × 105 cells/ml and incubated for 72h at 37°C. FMCA was performed using an automated Optimized Robot for Chemical Analysis (ORCA, Beckman Coulter, Fullerton, CA) programmed through the software SAMI (Beckman Coulter). The plates were washed, fluorescein diacetate (Sigma-Aldrich Sweden AB, Stockholm, Sweden) was added, and the fluorescence generated was measured at 485/520 nm using a fluorometer (Fluorostar Optima, BMG Technologies, Germany) after 50 min incubation. IC50 values were obtained from sigmoidal dose-response curves obtained using nonlinear regression of the survival plots for each drug and cell line, using GraphPad Prism (GraphPad Software Inc., La Jolla, CA, USA). All concentrations were tested in duplicates, and the experiments were repeated at least twice.
Quantitative real-time RT-PCR
RNA was prepared and analysed by quantitative real-time RT-PCR, as previously described for EGFR [Citation16], LRIG1 and 18S rRNA [Citation26], LRIG2 [Citation7] and LRIG3 [Citation27]. Triplicate samples of 20 ng of total RNA from exponentially growing cells were analysed, and relative RNA quantification was performed by comparing the threshold cycle values for the samples with standard curves generated with plasmid DNA containing cloned cDNA-fragments of respective genes. The mRNA levels were normalised to the 18S rRNA levels in respective samples.
Statistics
The survival functions were estimated with the Kaplan-Meier product limit estimator method and the median survival time estimated with linear interpolation of the survival function. The univariate statistical comparisons of the data were made using log-rank tests. Correlations to sensitivity for chemotherapeutic agents were analysed using Spearman rank order correlations. Kruskal-Wallis correlation test was used to determine statistical significance. Throughout this study, a 5% significance level was used in the statistical tests.
Results
Immunohistochemistry and clinical data
In the present study, a total of 80 patients were included. The most frequent histology was squamous cell carcinoma; further descriptive statistics are shown in . Protein expression analyses using an immunohistochemical method demonstrated widespread but variable expression levels of EGFR and LRIG1-3 in the tumours. In all cases a predominant cytoplasmic staining was observed (). Correlation analysis between immunohistochemically investigated proteins showed a significant correlation between EGFR and LRIG3 expression (p = 0.006, R = 0.37). The other investigated parameters were not significantly correlated to each other or to clinical parameters. Survival analysis showed that the total score, based on low or high expression of fraction and intensity, was not significantly correlated to survival for LRIG1 (p = 0.65) nor LRIG3 (p = 0.75). A trend towards decreased survival was found for low expression of EGFR (p = 0.09) () and high expression of LRIG2 (p = 0.18) (), although not statistically significant. Multivariate analyses of all investigated parameters in correlation to clinical data are shown in .
Figure 1. EGFR and LRIG1-3 expressed in tumours of human oesophageal cancer using an immunohistochemical staining procedure. Representative stainings of tumour tissues are shown. a: EGFR, b: LRIG1, c: LRIG2, d: LRIG3. Stainings were visualised at 40×magnification.
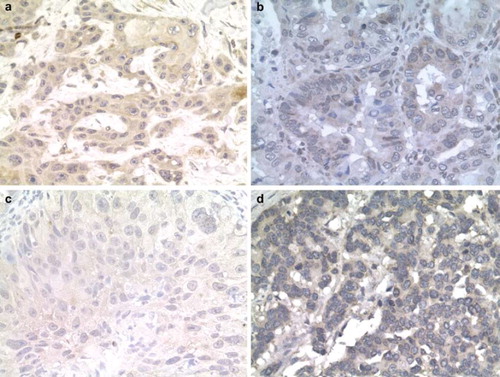
Figure 2a. Kaplan-Meier curves demonstrating the survival differences for patients with different tumour EGFR staining fraction and intensity, graded low or high. b. Kaplan-Meier curve demonstrating the survival differences for patients with different tumour LRIG2 staining fraction and intensity, graded low or high.
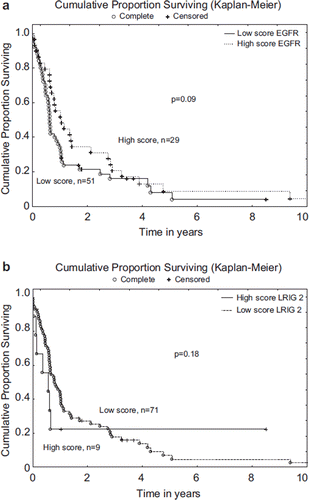
Figure 3a–c. Spearman rank order correlations describing significant correlations between expression levels of LRIG1 and chemosensitivity. a. Vinorelbine, R = −0.82 (p = 0.007) b. Docetaxel, R = −0.69 (p = 0.04) c. Cisplatin, R = −0.74 (p = 0.02).
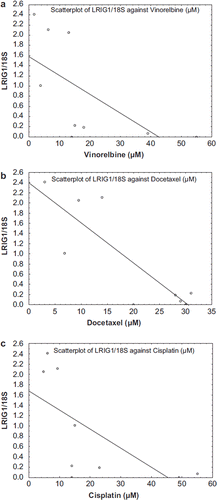
Table I. Description of patient characteristics and survival. Between 1990 and 2000, 126 patients were recorded as having received treatment for oesophageal carcinoma at the Department of Oncology, Uppsala University Hospital, Sweden. Formalin-fixed and paraffin-embedded tumour samples were obtained for immunohistochemical analysis of EGFR and LRIG1-3. The expression (staining extent) of EGFR and LRIG1-3 is expressed as the % of respective immunohistochemical staining score for each category (row) of the variables analysed.
Cytotoxicity assays and RT-PCR
The possible influence of EGFR family and LRIG proteins on sensitivity to conventional chemotherapeutic agents in a panel of oesophageal carcinoma cell lines was investigated. The RNA expression levels of ERBB1 (EGFR) and LRIG1-3 were evaluated by quantitative real-time RT-PCR and the sensitivity to a panel of cytotoxic drugs was determined by the FMCA method ( and ). Statistically significant correlations were found between expression of LRIG1 and sensitivity to cisplatin (r = −0.74), docetaxel (r = −0.69), and vinorelbine (r = −0.82) (, ).
Table II. The RNA expression levels of ERBB1(EGFR) and LRIG1-3 evaluated by quantitative RT-PCR. The mRNA levels were normalised to the 18S rRNA levels in respective samples.
Table III. IC50 values for seven standard cytotoxic drugs for nine ESCC cell lines using the fluorometric microculture cytotoxicity assay (FMCA). All concentrations were tested in duplicates, and the experiments were repeated at least twice. nd = not determined, could not be achieved by experimental conditions or analysed with nonlinear regression curve fit.
Table IV. Spearman rank order correlations between RT-PCR data on indicated receptors and chemosensitivity in vitro. All concentrations were tested in duplicates, and the experiments were repeated at least twice. Marked correlations (bold) are significant at p < 0.05.
Discussion
LRIG proteins have been proposed to be of importance in the pathogenesis of various tumours, [Citation11,Citation23] and LRIG1 has been shown to inhibit growth factor signalling from the oncogenic receptor tyrosine kinases EGFR, MET, and RET [Citation12–14,Citation28]. We evaluated for the first time the expression of LRIG proteins in oesophageal cancer. Our results show that there is a positive correlation between EGFR expression and LRIG3 expression. This is in contrast with data from Guo et al. [Citation29] who found that the expression of LRIG1-3 in the human pituitary adenoma cell line HP75 cell line was lower, but the expression of EGFR was higher, consistent with the notion of LRIG1-3 being tumour suppressor genes and that LRIGs decrease the expression of EGFR. Survival analysis showed no significant correlation to survival for LRIG1 (p = 0.65) or LRIG3 (p = 0.75). A trend towards decreased survival was found for low expression of EGFR (p = 0.09) () and high expression of LRIG2 (p = 0.18) (), although not statistically significant. This mirrors to some extent the situation in cervical squamous cell carcinoma, where high expression of LRIG2 is associated with poor survival [Citation20], whereas LRIG1 is associated with better survival [Citation18], indicating that the functions of LRIG1 and LRIG2 may be different. Furthermore, cytoplasmic LRIG2 expression was found to be an independent prognostic factor associated with poor oligodendroglioma patient survival [Citation19].
The incidence of EGFR-expressing oesophageal tumours remains controversial, with frequencies from as many as 99% to as few as 29% reported [Citation30]. Wang et al. reported that patients with oesophageal and oesophagogastric junction adenocarcinomas had shorter survival if EGFR expression was present, compared to those who were negative for EGFR [Citation31]. Furthermore, EGFR overexpression was significantly related to vascular invasion in squamous cell carcinoma patients. However, others have found the prognostic value of EGFR to be limited [Citation32,Citation33], and the function of EGFR protein expression as a prognostic factor is thus unclear. However, the EGFR pathway has been implicated in the pathophysiology of oesophageal cancer and EGFR inhibitors are being explored in these patients [Citation34]. The present study supports the notion that EGFR protein expression has limited prognostic implications in oesophageal carcinoma. Instead, other mechanisms may be involved in determining prognosis as indicated by Kaneko et al. who found that a silent mutation, comprised of a single nucleotide polymorphism (SNP) at codon 787 of exon 20 of the EGFR gene, was a negative prognostic factor in patients with esophageal squamous cell carcinoma [Citation33]. Furthermore, our analysis demonstrated no correlation between EGFR expression and tumour stage, as opposed to data by Wang et al. demonstrating that EGFR expression in oesophageal adenocarcinomas was correlated with advanced pathologic tumour classification and lymph node metastasis. In the same study EGFR expression was also correlated with poor disease-free and overall survival, but that correlation was not independent of T classification [Citation35].
The importance of the expression levels of EGFR and LRIG family members for the sensitivity of oesophageal carcinoma cell lines to conventional chemotherapeutic agents was investigated. Statistically significant correlations were found between expression levels of LRIG1 and sensitivity to cisplatin, docetaxel, and vinorelbine. The correlation between LRIG1 levels and sensitivity to cisplatin is consistent with reported results on malignant glioma cells, where ectopic expression of LRIG1 in U87MG EGFRvIII cells resulted in increased sensitivity to cisplatin and temozolomide [Citation36]. Also for targeted therapies, a possible link between LRIG1 activity and the sensitivity to EGFR tyrosine kinase inhibitors may be inferred, firstly because LRIG1 negatively regulates EGFR itself, and secondly because LRIG1 negatively regulates MET and ERBB3, which have been shown to mediate resistance to EGFR inhibition [Citation37]. Furthermore, there is evidence that EGFR signalling may confer resistance also to conventional chemotherapy. Thus, it seems possible that the effects of LRIG1 on the cellular sensitivity to cisplatin, docetaxel, and vinorelbine reported here and elsewhere is mediated, at least in part, by its suppression of EGFR signalling.
In summary, we show in the present study that EGFR and the LRIG proteins are widely expressed in oesophageal carcinoma, and may be functionally involved and of predictive but limited prognostic significance in oesophageal carcinoma. Further and larger studies are needed, however, to fully elucidate the prognostic and predictive value of EGFR and LRIG proteins in oesophageal carcinoma, and to reveal the functional relationships between these proteins and the aetiology of the disease and its response to therapy.
Acknowledgements
The authors would like to express their gratitude to Beatrice Tinge, M.D., Martin Dreilich, M.D., Ph.D., Kerstin Bergh, and Annika Holmberg for helpful assistance. The authors would like to express their gratitude for the financial support from the Cancer Foundation at Gavle Hospital, The Research Fund at the Department of Oncology, Uppsala University Hospital, the Cancer Research Foundation in Northern Sweden, the Swedish Research Council, and the Swedish Cancer Society. The authors declare that they have no competing interests.
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Devesa SS, Blot WJ, Fraumeni JF, Jr. Changing patterns in the incidence of esophageal and gastric carcinoma in the United States. Cancer 1998;83:2049–53.
- Law SY, Fok M, Wong J. Pattern of recurrence after oesophageal resection for cancer: Clinical implications. Br J Surg 1996;83:107–11.
- John MJ, Flam MS, Mowry PA, Podolsky WJ, Xavier AM, Wittlinger PS, . Radiotherapy alone and chemoradiation for nonmetastatic esophageal carcinoma. A critical review of chemoradiation. Cancer 1989;63:2397–403.
- Normanno N, Bianco C, Strizzi L, Mancino M, Maiello MR, De Luca A, Caponigro F, . The ErbB receptors and their ligands in cancer: An overview. Curr Drug Targets 2005;6: 243–57.
- Jankowski J, Hopwood D, Wormsley KG. Expression of epidermal growth factor, transforming growth factor alpha and their receptor in gastro-oesophageal diseases. Dig Dis 1993;11: 1–11.
- Teraishi F, Kagawa S, Watanabe T, Tango Y, Kawashima T, Umeoka T, . ZD1839 (Gefitinib, ‘Iressa’), an epidermal growth factor receptor-tyrosine kinase inhibitor, enhances the anti-cancer effects of TRAIL in human esophageal squamous cell carcinoma. FEBS Lett 2005;579:4069–75.
- Holmlund C, Nilsson J, Guo D, Starefeldt A, Golovleva I, Henriksson R, . Characterization and tissue-specific expression of human LRIG2. Gene 2004;332:35–43.
- Nilsson J, Starefeldt A, Henriksson R, Hedman H. LRIG1 protein in human cells and tissues. Cell Tissue Res 2003; 312:65–71.
- Guo D, Nilsson J, Haapasalo H, Raheem O, Bergenheim T, Hedman H, Henriksson R. Perinuclear leucine-rich repeats and immunoglobulin-like domain proteins (LRIG1-3) as prognostic indicators in astrocytic tumors. Acta Neuropathol 2006;111:238–46.
- Suzuki Y, Sato N, Tohyama M, Wanaka A, Takagi T. cDNA cloning of a novel membrane glycoprotein that is expressed specifically in glial cells in the mouse brain. J Biol Chem 1996;271:22522–7.
- Hedman H, Henriksson R. LRIG inhibitors of growth factor signalling – double-edged swords in human cancer? Eur J Cancer 2007;43:676–82.
- Laederich MB, Funes-Duran M, Yen L, Ingalla E, Wu X, Carraway KL, . The leucine-rich repeat protein LRIG1 is a negative regulator of ErbB family receptor tyrosine kinases. J Biol Chem 2004;279:47050–6.
- Gur G, Rubin C, Katz M, Amit I, Citri A, Nilsson J, Amariglio N, . LRIG1 restricts growth factor signaling by enhancing receptor ubiquitylation and degradation. EMBO J 2004;16: 3270–81. Epub 2004 Jul 3229.
- Shattuck DL, Miller J, Laederich M, Funes M, Petersen H, Carraway KL 3rd, Sweeney C. LRIG1 is a novel negative regulator of the Met receptor and opposes Met and Her2 synergy. Mol Cell Biol 2007;5:1934–46. Epub 2006 Dec 1918.
- Ledda F, Bieraugel O, Fard SS, Vilar M, Paratcha G. Lrig1 is an endogenous inhibitor of Ret receptor tyrosine kinase activation, downstream signaling, and biological responses to GDNF. J Neurosci 2008;28:39–49.
- Thomasson M, Hedman H, Guo D, Ljungberg B, Henriksson R. LRIG1 and epidermal growth factor receptor in renal cell carcinoma: A quantitative RT-PCR and immunohistochemical analysis. Br J Cancer 2003;89:1285–9.
- Tanemura A, Nagasawa T, Inui S, Itami S. LRIG-1 provides a novel prognostic predictor in squamous cell carcinoma of the skin: Immunohistochemical analysis for 38 cases. Dermatol Surg 2005;31:423–30.
- Lindstrom AK, Ekman K, Stendahl U, Tot T, Henriksson R, Hedman H, . LRIG1 and squamous epithelial uterine cervical cancer: Correlation to prognosis, other tumor markers, sex steroid hormones, and smoking. Int J Gynecol Cancer 2008;18:312–7.
- Holmlund C, Haapasalo H, Yi W, Raheem O, Brännström T, Bragge H, . Cytoplasmic LRIG2 expression is associated with poor oligodendroglioma patient survival. Neuropathology 2009;29:242–7.
- Hedman H, Lindström AK, Tot T, Stendahl U, Henriksson R, Hellberg D. LRIG2 in contrast to LRIG1 predicts poor survival in early-stage squamous cell carcinoma of the uterine cervix. Acta Oncol 49:812–5.
- Thomasson M, Hedman H, Guo D, Ljungberg B, Henriksson R. LRIG1 and epidermal growth factor receptor in renal cell carcinoma: A quantitative RT-PCR and immunohistochemical analysis. Br J Cancer 2003;89:1285–9.
- Holmlund C, Nilsson J, Guo D, Starefeldt A, Golovleva I, Henriksson R, Hedman H. Characterization and tissue-specific expression of human LRIG2. Gene 2004;332: 35–43.
- Guo D, Nilsson J, Haapasalo H, Raheem O, Bergenheim T, Hedman H, . Perinuclear leucine-rich repeats and immunoglobulin-like domain proteins (LRIG1-3) as prognostic indicators in astrocytic tumors. Acta Neuropathol 2006;111: 238–46.
- Bjorling E, Lindskog C, Oksvold P, Linne J, Kampf C, Hober S, . A web-based tool for in silico biomarker discovery based on tissue-specific protein profiles in normal and cancer tissues. Mol Cell Proteomics 2008;7:825–44.
- Nygren P, Fridborg H, Csoka K, Sundström C, de la Torre M, Kristensen J, Bergh J, . Detection of tumor-specific cytotoxic drug activity in vitro using the fluorometric microculture cytotoxicity assay and primary cultures of tumor cells from patients. Int J Cancer 1994;5:715–20.
- Nilsson J, Vallbo C, Guo D, Golovleva I, Hallberg B, Henriksson R, . Cloning, characterization, and expression of human LIG1. Biochem Biophys Res Commun 2001;284:1155–61.
- Guo D, Holmlund C, Henriksson R, Hedman H. The LRIG gene family has three vertebrate paralogs widely expressed in human and mouse tissues and a homolog in Ascidiacea. Genomics 2004;84:157–65.
- Ledda F, Bieraugel O, Fard SS, Vilar M, Paratcha G. Lrig1 is an endogenous inhibitor of ret receptor tyrosine kinase activation, downstream signaling, and biological responses to GDNF. J Neurosci 2008;28:39–49.
- Guo D, Han L, Shu K, Chen J, Lei T. Down-regulation of leucine-rich repeats and immunoglobulin-like domain proteins (LRIG1-3) in HP75 pituitary adenoma cell line. J Huazhong Univ Sci Technolog–Med Sci 2007;27:91–4.
- Kii T, Takiuchi H, Kawabe S, Gotoh M, Ohta S, Tanaka T, . Evaluation of prognostic factors of esophageal squamous cell carcinoma (stage II III) after concurrent chemoradiotherapy using biopsy specimens. Jpn J Clin Oncol 2007; 37:583–9.
- Kim LW, Tsung-Teh W, In Seon C, Huamin W, Erika R, Arlene MC, . Expression of epidermal growth factor receptor in esophageal and esophagogastric junction adenocarcinomas. Cancer 2007;109:658–67.
- Wang L-S, Chow K-C, Chi K-H, Liu C-C, Li W-Y, Chiu J-H, . Prognosis of esophageal squamous cell carcinoma: Analysis of clinicopathological and biological factors. Am J Gastroenterol 1999;94:1933–40.
- Kaneko K, Kumekawa Y, Makino R, Nozawa H, Hirayama Y, Kogo M, Konishi K, . EGFR gene alterations as a prognostic biomarker in advanced esophageal squamous cell carcinoma. Front Biosci 2010;15:65–72.
- Campen TDaC. Anti-EGFR-targeted therapy for esophageal and gastric cancers: An evolving concept. J Oncol 2009;2009.
- Wang KL, Wu T-T, Choi IS, Wang H, Resetkova E, Correa AM, . Expression of epidermal growth factor receptor in esophageal and esophagogastric junction adenocarcinomas. Cancer 2007;109:658–67.
- Stutz MA, Shattuck DL, Laederich MB, Carraway KL, 3rd, Sweeney C. LRIG1 negatively regulates the oncogenic EGF receptor mutant EGFRvIII. Oncogene 2008;27:5741–52.
- Nguyen KS, Kobayashi S, Costa DB. Acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancers dependent on the epidermal growth factor receptor pathway. Clin Lung Cancer 2009;10:281–9.
