Abstract
Introduction. Somatic mutations in plant homeodomain finger protein 6 (PHF6) gene have recently been reported in T-cell acute lymphoblastic leukemia (T-ALL), strongly suggesting its role in the pathogenesis of human cancers. Materials and methods. To see whether the PHF6 mutation occurs in other malignancies, we analyzed entire coding sequences of PHF6 in 231 hematologic malignancies [105 acute myelogenous leukemias (AML), 66 pre-B-ALL, 23 T-ALL, one undifferentiated acute leukemia and 36 multiple myelomas] by single-strand conformation polymorphism assay. Also, we analyzed the mutation in 236 solid cancers, including 41 lung, 39 hepatocellular (HCC), 36 breast, 40 colorectal, 40 gastric and 40 prostate carcinomas. Results. In the hematologic malignancies, there were 11 PHF6 mutations that were detected not only in T-ALL (34.7%) (five adult and three childhood T-ALL), but also in two AML (1.9%) (one acute monocytic leukemia and one AML minimally differentiated). In addition, there was a PHF6 mutation in the HCC (2.6%). The PHF6 mutations were detected in both male and female patients, and consisted of six frameshift, three nonsense and two intron mutations. Conclusion. Our data suggest that PHF6 mutation might play a role in tumorigenesis not only of T-ALL, but also of AML and HCC.
Plant homeodomain finger protein 6 (PHF6) gene encodes a protein with two PHD-type zinc finger domains [Citation1]. PHF6 protein is localized in nucleus and nucleolus, and is considered a potential transcription factor [Citation2]. PHF6 gene localized to human chromosome Xq26.3 and is mutated in the germline of Börjeson-Forssman-Lehmann syndrome that is a rare X-linked mental retardation syndrome [Citation2]. PHF6 is involved in mitosis and DNA damage response [Citation3–5], but its other cellular functions remain largely unknown.
Several lines of evidence indicate that PHF6 gene is implicated in leukemogenesis [Citation6]. PHF6 gene mutation has recently been identified in sporadic T-cell acute lymphoblastic leukemia (T-ALL) [Citation7]. T-ALL has an increased incidence in males, and in the course of identifying the X-linked genes in T-ALL, Vlierberghe et al. [Citation7] found that significant numbers of adult (38%) and childhood (16%) T-ALL (total 22.9%) harbored PHF6 mutation. The PHF6 mutations consisted of nonsense and frameshift mutations (70%), and missense mutations (30%), suggesting that most of the mutations might inactivate the functions of PHF6 . A following study found that PHF6 gene is mutated in acute myelogenous leukemias (AML) at a lower frequency (2.8%) [Citation8]. These reports showed that PHF6 gene might be a tumor suppressor gene in human.
One of the main concerns in cancer genetics is to address whether any cancer-causing mutation is specific to few cancer types or widespread. For example, EGFR and JAK2 mutation is specific to few cancer types [Citation9,Citation10], whereas K-RAS and TP53 mutations are common to many cancer types [Citation11,Citation12]. Because PHF6 is ubiquitously expressed in tissues [Citation7], it could be hypothesized that alterations of PHF6 gene could be responsible to the pathogenesis of other cancers as well as T-ALL. To date, however, the data on the mutation status of PHF6 gene is available only in T-ALL and AML. In order to further characterize PHF6 mutation in human cancers, we investigated whether human cancer tissues from various histologic origins harbor PHF6 mutations.
Material and methods
Tissue samples and microdissection
The tissue specimens consisted of 231 hematologic and 236 solid malignancies. Acute leukemia (n = 195) and multiple myeloma (n = 36) DNA samples were extracted from the bone marrow of 204 hematologic malignancies. The DNA samples from the same patients were extracted from the bone marrows after complete remission if available, and used as possible normal genomic DNA samples of the same patients. Acute leukemias consisted of 105 AML, 89 ALL (66 pre-B-ALL and 23 T-ALL) and one undifferentiated acute leukemia. All of the AML and 30 pre-B-ALL were adult leukemias, while 36 pre-B-ALL were childhood pre-B-ALL. The T-ALL consisted of 14 adult and nine childhood T-ALL. The AML samples consisted of eight AMLs minimally differentiated (FAB classification: M0), 15 AMLs without maturation (M1), 21 AMLs with maturation (M2), 15 AML with t(8;21)(q22;q22) (M2), 12 acute promyelocytic leukemias (AML with t(15;17)(q22;q12)) (M3), seven acute myelomonocytic leukemias (M4), six AMLs with abnormal bone marrow eosinophils Inv(16)(p13q22) (M4), seven acute monoblastic and monocytic leukemias (M5), two acute erythroid leukemias (M6), 11 AMLs with multilineage dysplasia and one AML and myelodysplastic syndrome, therapy-related according to the WHO classification.
Methacarn-fixed solid tissues of 41 non-small cell lung cancers, 39 hepatocellular carcinomas (HCC), 36 breast carcinomas, 40 prostate carcinomas, 40 colorectal carcinomas and 40 gastric carcinomas were randomly selected for this study. The lung cancers consisted of 22 adenocarcinomas and 19 squamous cell carcinomas. The breast carcinomas consisted of 36 invasive ductal carcinomas. The colorectal carcinomas originated from cecum (n = 2), ascending colon (n = 6), transverse colon (n = 1), descending colon (n = 1), sigmoid colon (n = 8) and rectum (n = 24). The gastric carcinomas consisted of 21 diffuse-type and 19 intestinal-type by Lauren's classification, and eight early and 32 advanced gastric carcinomas according to depth of invasion. All of the patients were Asians (Korean). Approval was obtained from the Catholic University of Korea, College of Medicine's institutional review board for this study. We analyzed the primary tumors, but not the metastatic lesions. We did not include the cancer cell lines in this study. For the solid tumors, tumor cells and normal cells from the same patients were selectively procured from hematoxylin and eosin-stained slides using a 30G1/2 hypodermic needle (Becton Dickinson, Franklin Lakes, NJ) affixed to a micromanipulator by the microdissection, as described previously [Citation13,Citation14]. DNA extraction was performed by a modified single-step DNA extraction method by proteinase K treatment.
Single strand conformation polymorphism (SSCP) analysis for mutation detection
In the previous report, PHF6 gene mutations in T-ALL were detected widely in the coding sequences [Citation7]. Thus, we analyzed PHF6 mutations in the entire coding region. Genomic DNA each from tumor cells and normal cells were amplified with nine primer pairs covering the entire coding exons (exon 2-10) (). Radioisotope ([32P]dCTP) was incorporated into the PCR products for detection by autoradiogram. The polymerase chain reaction (PCR) reaction mixture was denatured for 1 min at 94oC and incubated for 30 cycles (denaturing for 30 s at 94oC, annealing for 30 s at 50oC, and extending for 30 s at 72oC). After amplification, PCR products were denatured and were loaded onto SSCP gel (Cambrex Bio Science Rockland, Rockland, ME, USA) with 10% glycerol. After overnight SSCP at a room temperature, the gels were transferred to 3-mm Whatman paper and dried, and autoradiography was performed. Other procedures of the PCR-SSCP were described in our previous studies [Citation13,Citation14]. Migration of the PCR products on SSCP was analyzed by visual inspection. After SSCP, DNAs showing mobility shifts were cut out from the dried gel, and re-amplified for 30 cycles using the same primer sets for DNA sequencing. Sequencing of the PCR products was carried out using a capillary automatic sequencer. To confirm the SSCP data, we repeated the PCR-SSCP twice. In the second round SSCP, we included positive controls of the mutations that had been detected in the first round SSCP.
Table I. Primer sequences used for PCR amplification of individual exons of PHF6.
Results and discussion
First, we analyzed the PHF6 mutation in the hematologic malignancies. PCR and subsequent SSCP analysis identified 10 aberrantly migrating bands compared to wild-type bands (). None of the corresponding normal samples from the same patients showed evidence of mutations by SSCP (), indicating the mutations had risen somatically. Enrichment and DNA sequence analysis of the aberrantly migrating bands of the SSCP led us to identify that 10 hematologic malignancies harbored PHF6 mutations ( and ). The mutations were detected in T-ALL (8/23; 34.7%) and AML (2/105; 1.9%), but neither in pre-B-ALL or multiple myelomas. The two AML were an acute momocytic leukemia and an AML minimally differentiated.
Figure 1. Representative SSCP and DNA sequencing of PHF6 gene in the cancers. SSCP (A–C) and DNA sequencing analysis (D–F) of PHF6 gene from a T-ALL, an AML and a HCC in cancer (Lane T) and normal tissues (Lane N). A–C. The arrows in lane T indicate aberrant bands as compared with SSCP of normal tissues (lane N). D–F. DNA sequencing of the aberrant SSCP bands in A, B and C show four base insertions in D (c.90_91insCCCG), one base duplication in E (c.27dupA) and a substitution of a base in F (c.673C > T), respectively.
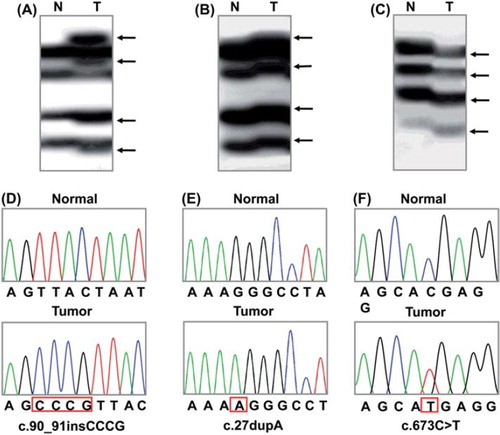
Table II. Summary of PHF6 mutations.
Genomic DNAs isolated from normal and tumor tissues of 41 non-small cell lung cancers, 39 HCC, 36 breast carcinomas, 40 prostate carcinomas, 40 colorectal carcinomas and 40 gastric carcinomas were also analyzed for the detection of PHF6 mutations. By the PCR-SSCP, we found one PHF6 mutation in HCC (1/39; 2.6%). SSCP from other cancers did not reveal any aberrantly migrating band compared to the wild-type band from the normal tissues, indicating there was evidence of PHF6 mutation. We repeated the experiments twice, including PCR, SSCP and DNA sequencing analysis to ensure the specificity of the results, and found that the data were consistent (data not shown).
The PHF6 mutations consisted of six frameshift mutations, three nonsense mutation and two mutations in introns. With respect to ages, the mutations were detected in both adult T-ALL (5/14; 35.7%) and childhood T-ALL (3/9; 33.3%) (). All of the patients with AML and HCC harboring PHF6 mutations were adults. With respect to genders, the mutations were detected in both male T-ALL (6/17; 35.3%) and female T-ALL (2/6; 33.3%) (). In AML and HCC, PHF6 mutations were found in male cancers (2/59; 3.4% and 1/25; 4.0%, respectively), but not in female cancers (0/46 and 0/14, respectively). All of the patients with AML and HCC harboring PHF6 mutations were males. Neither age or gender was not associated with the PHF6 mutation incidence (χ2 test, p < 0.05). The mutation data are summarized in .
Discovery of PHF6 mutation in T-ALL raised several critical questions. One of these was whether other malignancies besides T-ALL harbor PHF6 somatic mutation, because PHF6 protein is ubiquitously expressed in human tissues and is considered a tumor suppressor. In this study, we attempted to find whether the PHF6 somatic mutation is present in hematologic malignancies as well as in common solid cancers. We found that T-ALL (8/23; 34.7%) and AML (2/105; 1.9%) harbored PHF6 mutations. These results are similar to the previous ones by other researchers (T-ALL: 22.9%, AML: 2.9%) [Citation7,Citation8], and there was no significant difference between our data and those data (Fisher's exact test, p > 0.05). Also, most of the PHF6 mutations (81.8%) detected in our study were frameshift or nonsense mutations, which were also common in the previous studies [Citation7,Citation8]. Above data suggest that PHF6 somatic mutation occurs in T-ALL and AML in both western [Citation7,Citation8] and Asian (Korean in our study) populations.
The two earlier studies reporting PHF6 mutations revealed that the mutations statistically more common in male patients [Citation7,Citation8]. In the T-ALL, the mutations are about 12 times more common in male (31.5%) than in female (2.6%) [Citation7]. In the AML, they are seven times more common in male (4.6%) than in female (0.6%) [Citation8]. However, in our data PHF6 mutation incidences between male and female patients of T-ALL or AML or HCC were not significantly different (Fisher's exact test, p > 0.05). Even if all of them are added together, they do not show any significant differences (9/101 vs. 2/66). It is unclear that the discrepancy was originated from ethnicity or statistical problems. For these points, studies are now needed that attempt to find PHF6 mutations in other countries.
To our knowledge, there have been no mutational studies on PHF6 gene in solid cancers. In the present study, we screened PHF6 mutation in common solid cancers from liver, stomach, colon, prostate, lung and breast. We found one mutation in HCC (2.6%). Despite the low incidence in HCC, the inactivating nature of the mutation (nonsense mutation) suggests that the mutation may not be a passenger mutation. By contrast, other solid cancers were negative for PHF6 mutation, indicating that PHF6 mutation may not play a crucial role in development of solid cancers and suggest that PHF6 mutations may be specific to hematopoietic malignancies, especially T-ALL. However, it remains to be clarified that other tumors, including mesenchymal, neuroendocrine, glial, lymphoid tumors, or melanomas are absent for the PHF6 mutation.
Acknowledgements
This study was supported by a grant from Ministry for Health, Welfare and Family Affairs (A100098).
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Baker LA, Allis CD, Wang GG. PHD fingers in human diseases: Disorders arising from misinterpreting epigenetic marks. Mutat Res 2008;647:3–12.
- Gécz J, Turner G, Nelson J, Partington M. The Börjeson-Forssman-Lehman syndrome (BFLS, MIM #301900). Eur J Hum Genet 2006;14:1233–7.
- Dephoure N, Zhou C, Villén J, Beausoleil SA, Bakalarski CE, Elledge SJ, . A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci USA 2008;105:10762–7.
- Lowndes NF, Toh GW. DNA repair: The importance of phosphorylating histone H2AX. Curr Biol 2005;15:R99–102.
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER 3rd, Hurov KE, Luo J, . ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 2007;316:1160–6.
- Landais S, Quantin R, Rassart E. Radiation leukemia virus common integration at the Kis2 locus: Simultaneous overexpression of a novel noncoding RNA and of the proximal Phf6 gene. J Virol 2005;79:11443–56.
- Van Vlierberghe P, Palomero T, Khiabanian H, Van der Meulen J, Castillo M, Van Roy N, . PHF6 mutations in T-cell acute lymphoblastic leukemia. Nat Genet 2010;42:338–42.
- Van Vlierberghe P, Patel J, Abdel-Wahab O, Lobry C, Hedvat CV, Balbin M, . PHF6 mutations in adult acute myeloid leukemia. Leukemia 2011;25:130–4.
- James C, Ugo V, Le Couedic JP, Staerk J, Delhommeau F, Lacout C, . A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature 2005;434:1144–8.
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, . Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004; 350:2129–39.
- Braithwaite AW, Prives CL. p53: More research and more questions. Cell Death Differ 2006;13:877–80.
- Malumbres M, Barbacid M. RAS oncogenes: The first 30 years. Nat Rev Cancer 2003;3:459–65.
- Lee JW, Soung YH, Kim SY, Lee HW, Park WS, Nam SW, . PIK3CA gene is frequently mutated in breast carcinomas and hepatocellular carcinomas. Oncogene 2005;24: 1477–80.
- Lee SH, Shin MS, Park WS, Kim SY, Kim HS, Han JY, . Alterations of Fas (Apo-1/CD95) gene in non-small cell lung cancer. Oncogene 1999;18:3754–60.
