Abstract
Objective
To safeguard key workers involved in development and production of medicines and ensure business continuity, we developed an occupational healthcare program, performed by our company’s occupational healthcare services, to assess the infection and immune status for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). This pilot program, conducted at our company facilities, evaluated the suitability of diagnostic tools in our setting for program upscaling.
Methods
We used different marketed in vitro diagnostics (including tests for antibodies against spike protein subunits S1 and S2 and nucleocapsid [N] protein) combined with medical history, symptoms and likelihood of infection. We evaluated the testing strategy over four visits in 141 employees (known positive COVID-19 history, n = 20; unknown status, n = 121) between April and June 2020 at four company locations in Germany. Digital self-monitoring over the pilot program duration was also included.
Results
No incident infections were detected. Based on immune status, medical history and likelihood of infection, 10 participants (8.3%) with previously unknown history of COVID-19 were identified to have been infected before entering the program. These participants, who recalled no or mild symptoms in the preceding months, were primarily identified using an assay that detected both S1 and S2 immunoglobulin (Ig) G. The frequency of positive lateral flow assay (LFA) results (IgM or IgG directed against the N-protein) in this cohort was lower compared with participants with a known history of COVID-19 (0‒10.8% vs. 33.8‒75.7%, respectively).
Conclusions
Data from this pilot program suggest that LFA for antibodies may not always reliably detect current, recent or past infections; consequently, these have not been included in our upscaled occupational healthcare program. Regular testing strategies for viral RNA and antibodies directed against different SARS-CoV-2 proteins, combined with hygiene rules and a comprehensive baseline assessment, are recommended to ensure avoidance of infections at workplace as reliably as possible.
Keywords:
Introduction
In response to the pandemic outbreak of COVID-19, governments across the world have recommended that employees work from home if they are able to do so; however, in some instances, physical presence needs to be maintained. Boehringer Ingelheim (BI) has several manufacturing plants in Germany that produce items on the World Health Organization’s list of essential medications, and therefore requires around 6,000 members of staff to stay onsite. In addition to hygiene measures such as mandatory mask wearing, “bubble shifts” and physical distance between workers, BI’s occupational healthcare services have launched an occupational healthcare program (“COVID-19 Testing@BI”) for detection and monitoring of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection and immune status. The aim of the program is to identify and, if necessary, quarantine infected staff as quickly and effectively as possible, thereby safeguarding the health of all employees and maintaining business continuity. Before commencing this large-scale program, BI carried out a pilot phase to establish the protocol and to evaluate a set of suitable diagnostic tools. The pilot phase started approximately 6 weeks after the first peak of the pandemic in Germany, in mid-March 2020. At this point in time, the SARS-CoV-2 infection incidence was declining due to extensive social distancing, hygiene and lockdown measures [Citation1]. It ended in early June 2020, when SARS-CoV-2 infection incidence was still at a relatively low level.
The current “gold standard” method used to identify an existing SARS-CoV-2 infection is by detection of viral RNA from nasopharyngeal/oropharyngeal swabs or sputum samples by reverse transcription polymerase chain reaction (RT-PCR). In addition, investigating SARS-CoV-2-specific antibodies helps to detect an occurred immune response. Most individuals infected with SARS-CoV-2 develop specific immunoglobulins (Igs; e.g. IgM, IgA or IgG) in response to the different proteins and epitopes of the virus, i.e. internal nucleocapsid (N) protein, and S1 and S2 subunits of the spike (S) glycoprotein [Citation2]. These can be detected with commercially available SARS-CoV-2-specific diagnostic tests, e.g. lateral flow assays (LFAs) or laboratory-based assays (e.g. enzyme-linked immunosorbent assays [ELISAs]). However, SARS-CoV-2-RNA and different antibodies against SARS-CoV-2 can be detected at different time points of the disease [Citation3], and there is a diagnostic “blind spot” during very early infection (Supplementary Figure S1) when infection may be undetected. Initial false-negative RT-PCR test results in pre-symptomatic patients make it difficult to contain viral spread [Citation4]. For effective workforce monitoring, an assessment of medical history, symptoms and likelihood of infection can help to provide a more comprehensive picture.
The longevity of antibody response and resulting immunity against SARS-CoV-2 is not yet clear: some studies report that the IgG response is relatively stable over time [Citation5,Citation6], whereas others found diminution of IgG antibody titers, particularly when single Ig assays were employed (as opposed to pan-Ig assays that measure IgA, IgM and IgG) [Citation5,Citation7]. A stronger antibody response has been described in patients with severe COVID-19 compared with asymptomatic or mildly ill individuals [Citation2]. This pattern of antibody response may be of relevance to the clinical course of the disease; a recent study reports higher ratios of IgG antibodies targeting S1 or receptor-binding domains of spike compared with N antigens in outpatients with mild illness versus severely ill patients [Citation2].
Here, we describe the design and results of the pilot phase of COVID-19 Testing@BI. The objective of this pilot study was to evaluate the suitability of different diagnostic methods (detection of viral RNA, immune response to different SARS-CoV-2 proteins, and a comprehensive assessment of medical history, clinical symptoms and likelihood of infection over time) in a group of BI employees.
Methods and materials
Study design and participants
The pilot program took place from April‒June 2020 and included employees aged between 20 and 63 years, from four BI sites in Germany. Voluntary participation could be withdrawn at any point. Participants provided signed/dated written consent. Ethical approval was obtained from the Rhineland-Palatinate Medical Association prior to data analyses. Since COVID-19 Testing@BI is an occupational healthcare program and not a clinical study, regulatory requirements of clinical development purposes were not applied. For more information on data protection, see Supplementary Appendix: Methods.
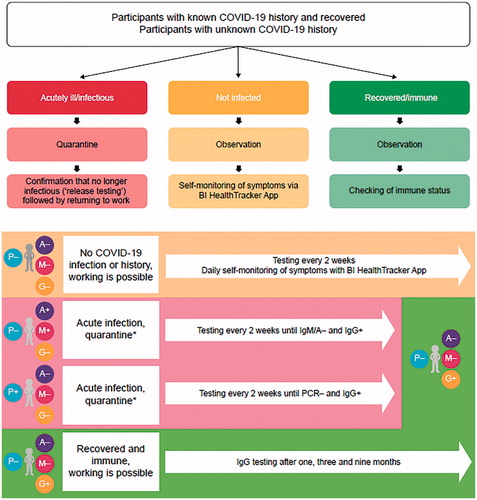
The pilot was performed as a two-group testing program according to COVID-19 history: (i) known and recovered from COVID-19 and (ii) unknown COVID-19 history. This design enabled comparison of test results between a group known to have had COVID-19 (COVID-19 known positive [COVID-19kp]) and a group with unknown history, but potentially infected unknowingly in the past (COVID-19uk). Testing schedule and subsequent actions were determined by participants’ status, which included not infected, acutely ill/infectious, or recovered/immune ( and Supplementary Table S1). Four regular visits were scheduled at approximately 2-week intervals (see Supplementary Appendix).
Figure 1. Participant categories at the time of entering the program and testing schedule. *While in quarantine (due to COVID-19 infection), testing days are postponed until official release of quarantine. In Germany, only the respective health authority can send into quarantine and discharge from quarantine. Abbreviations. A, immunoglobulin A; G, immunoglobulin G; M, immunoglobulin M; P, reverse-transcription real-time PCR; PCR, polymerase chain reaction.
Baseline and disease characteristics
A comprehensive assessment of participants’ baseline and disease characteristics including medication use and influenza vaccination was performed.
Testing materials
All test kits were CE-marked and commercially available.
For detection of SARS-CoV-2 RNA by RT-PCR, an oropharyngeal swab was collected by qualified healthcare personnel. Samples were assayed at BI sites at Biberach and Hanover using the Kylt SARS-CoV-2 Initial Screening/Confirmation (AniCon Labor GmbH, Höltinghausen, Germany). Initiation of a confirmatory RT-PCR by an accredited laboratory for positive initial RT-PCR results and IgM- (LFA) and IgA- (ELISA) positive participants was performed by a contracted partner, the accredited laboratory Bioscientia Healthcare GmbH, Ingelheim, Germany.
LFAs for IgM and IgG determination were used at BI sites using fingertip blood or venous blood samples. The following test systems were evaluated: VivaDiag COVID-19 IgM/IgG Rapid Test (VivaChek Biotech Co Ltd, Hangzhou, China); 2019-nCoV IgG/IgM Rapid Test Cassette (MöLab GmbH, Langenfeld, Germany); SARS-CoV-2 rapid IgM/IgG antibody test (Nano Repro AG, Marburg, Germany); and PerGrande SARS-CoV-2 Antibody Detection Kit IgG (PerGrande BioTech Development Co Ltd, Beijing, China); all directed against the N protein of the virus.
Laboratory assays for specific SARS-CoV-2 IgA, IgG and total IgE (unspecific) were conducted at Bioscientia using venous blood samples (IgG S1/2: LIAISON SARS-CoV-2 S1/S2 IgG chemiluminescence immunoassay [CLIA; Diasorin, Saluggia, Italy]; IgG S1: Anti-SARS-CoV-2 ELISA S1 IgG [Euroimmun AG, Lübeck, Germany]; IgA (S1): anti-SARS-CoV-2 ELISA IgA [Euroimmun AG]; total IgE: Roche Elecsys Total IgE II assay [Roche Diagnostics, Rotkreuz, Switzerland]). All laboratory assays outlined above allow a quantitative readout. This feature was used to capture the temporal evolution of the antibody levels. For company-provided information of sensitivity and specificity, please see Supplementary Table S2.
Digital self-monitoring
For real-time digital self-monitoring, employees were asked to record vital signs (body temperature and pulse) and any suspicious symptoms for COVID-19 daily on a HealthTracker App, developed by the BI IT department in collaboration with the medical team. App access could only be obtained by enrolled employees, via named user access control.
Statistical analysis
Test results and demographic data were analyzed using descriptive statistics. Standard statistical parameters of the test performance, like sensitivity, specificity, and positive and negative predictive values, were calculated (see Supplementary Appendix: Methods; Supplementary Figure S2). To that end, medical judgment and a post hoc classification of each participant’s COVID-19 history—based on the full diagnostic information package accumulated during the testing program—was considered as gold standard. The (semi-) quantitative outcomes from the laboratory assay tests were plotted over time and modeled using linear regression to quantify the dynamics of potential seroconversion.
Results
Participants and baseline characteristics
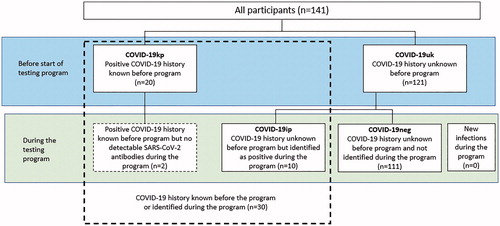
Baseline demographics and characteristics are shown in and Supplementary Table S3. In total, 141 employees aged between 20 and 63 years entered the program (median age 45 years; 61 female; 43.3%); amongst these, 20 participants were known positives for COVID-19 (COVID-19kp; confirmed by RT-PCR and clinical assessment). There were no incident infections during the testing phase (Supplementary Table S4). In two participants, RT-PCR results were still positive 4–6 weeks after the participants had been released from quarantine. Based on clinical judgment, it was decided to recommend prolongation of home-based isolation and re-testing until negative PCR before entering the site for work was allowed. A further two participants in the COVID-19kp cohort did not develop any measurable antibody response during the course of our study.
Table 1. COVID infection by demographics and baseline characteristics.
Ten participants (8.3%) out of the 121 COVID-19uk participants were medically judged to have had COVID-19 prior to testing, based on diagnostic results, evidence for risk of infection and medical history, indicating a mild course of infection with only minor clinical symptoms; this subset was subsequently denoted as COVID-19-identified-positive (COVID-19ip; Supplementary Table S5). Of these, nine participants were positive by Diasorin S1/S2 IgG CLIA (including two who were also positive for S1 IgA and one also positive for both S1 IgA/IgG). A further participant was positive by IgA/IgG S1 ELISA at Visit 1, but not S1/S2 IgG. The remaining 111 participants from the COVID-19uk cohort were re-classified as COVID-19 negative (COVID-19neg; and ). Participants from the COVID-19kp or COVID-19ip groups were numerically less likely to report allergies compared with the COVID-19neg group, including the most commonly reported allergy “allergic rhinitis due to pollen”, less likely to take antihistamines, to have baseline IgE values of ≥100 IU/mL and less likely to have had an influenza vaccination in 2019/2020 ( and Supplementary Figure S3).
Figure 2. COVID-19 infection status of participants before and during the pilot program. Abbreviations. COVID-19ip, COVID-19 history unknown before the program but identified as positive during the program; COVID-19kp, positive COVID-19 history known before program; COVID-19neg, COVID-19 history unknown before program and not identified during the program; COVID-19uk, COVID-19 history unknown before program; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Detection of SARS-CoV-2 antibodies by LFA
Full details of results by LFA and Ig subtype are given in Supplementary Tables S6 and S7. Amongst COVID-19kp participants, the frequency of participants with positive LFA IgG and IgM testing results generally decreased over time, except for the MöLab 2019-nCoV IgM LFA. In COVID-19ip participants, the frequency of participants with positive LFA results (IgM and IgG) was much lower compared with COVID-19kp participants (0‒10.8% vs. 33.8‒75.7%, respectively).
Positive IgM results in the absence of additional indication of SARS-CoV-2 infection were found in 10 (8.3%) COVID-19neg participants, using at least one LFA and at least once over the observation period. In most cases, these participants had an allergic disposition with symptoms of seasonal allergic reactions and/or potential infections during testing, with subacute symptoms (such as pain or increased body temperature) as reflected in the BI HealthTracker. None of the participants were RT-PCR positive and no other SARS-CoV-2 antibodies were observed at any visit. It is therefore likely that these isolated IgM results were unspecific and probably caused by infection other than SARS-CoV-2.
Detection of SARS-CoV-2 antibodies by laboratory serology
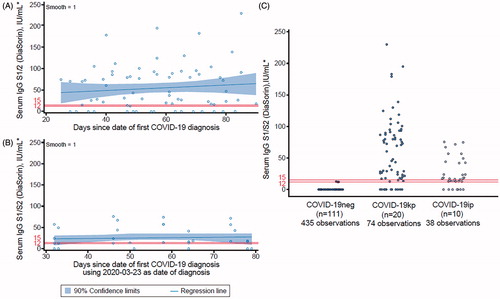
A pronounced decline in S1 IgA (Euroimmun) levels was observed over time, in particular amongst COVID-19kp participants (). In comparison, the overall assay levels of the COVID-19ip group () fell clearly below the COVID-19kp group, with levels that were hardly sufficient to generate a positive test signal (). Positive and negative test results for IgA against S1 protein in participants over time are shown in Supplementary Table S8.
Figure 3. IgA against S1 protein (Euroimmun) over time. Panel (A) Participants with COVID-19 history known before the program start (n = 20), slope per day (confidence intervals): –0.08 (–0.14, –0.02); (B) Participants with COVID-19 history identified during the program (n = 10); (C) Comparison of levels in all participants (n = 141) over all four visits. *An optical density [OD] ratio of serum sample/OD of calibrator below 0.8 was evaluated as negative, 0.8–<1.1 as borderline and >1.1 as positive (positive/negative thresholds denoted by red lines/horizontal lines in print). Linear regression (locally estimated scatterplot smoothing) including 90% confidence limits are indicated in shading. Maximum values shown were >13.0 in the raw data set and set to 13.0 for analysis. Date of positive RT-PCR for the COVID-19kp group, median (min, max): 2020 − 03 − 23 (2020 − 03 − 11, 2020 − 03 − 31). The same date was arbitrarily assumed for the COVID-19ip group (which also coincided with the first lockdown in Germany). Abbreviations. COVID-19ip, COVID-19 history unknown before program but identified as positive during the program; COVID-19kp, positive COVID-19 history known before program; COVID-19neg, COVID-19 history unknown before program and not identified during the program; IgA, immunoglobulin A; RT-PCR, reverse transcription polymerase chain reaction.
![Figure 3. IgA against S1 protein (Euroimmun) over time. Panel (A) Participants with COVID-19 history known before the program start (n = 20), slope per day (confidence intervals): –0.08 (–0.14, –0.02); (B) Participants with COVID-19 history identified during the program (n = 10); (C) Comparison of levels in all participants (n = 141) over all four visits. *An optical density [OD] ratio of serum sample/OD of calibrator below 0.8 was evaluated as negative, 0.8–<1.1 as borderline and >1.1 as positive (positive/negative thresholds denoted by red lines/horizontal lines in print). Linear regression (locally estimated scatterplot smoothing) including 90% confidence limits are indicated in shading. Maximum values shown were >13.0 in the raw data set and set to 13.0 for analysis. Date of positive RT-PCR for the COVID-19kp group, median (min, max): 2020 − 03 − 23 (2020 − 03 − 11, 2020 − 03 − 31). The same date was arbitrarily assumed for the COVID-19ip group (which also coincided with the first lockdown in Germany). Abbreviations. COVID-19ip, COVID-19 history unknown before program but identified as positive during the program; COVID-19kp, positive COVID-19 history known before program; COVID-19neg, COVID-19 history unknown before program and not identified during the program; IgA, immunoglobulin A; RT-PCR, reverse transcription polymerase chain reaction.](/cms/asset/6257fa63-d094-4c04-977b-c3e3d003000a/icmo_a_1914943_f0003_c.jpg)
A similar decline in levels was observed for the S1-specific IgG response (Euroimmun) in the COVID-19kp group (). As observed with IgA, the overall assay levels of the COVID-19ip group fell clearly below the “known positive” level () and were therefore not sufficient to generate a positive test signal (). Positive and negative test results for IgG against S1 protein in participants over time are shown in Supplementary Table S9.
Figure 4. IgG against S1 protein (Euroimmun) over time. Panel (A) Participants with COVID-19 history known before the program (n = 20), slope per day (confidence intervals): –0.08 (–0.12, –0.03); (B) Participants with COVID-19 history identified during the program (n = 10); (C) Comparison of levels in all participants (n = 141) over all four visits. *An optical density [OD] ratio of serum sample/OD of calibrator below 0.8 was evaluated as negative, 0.8–<1.1 as borderline and >1.1 as positive (positive/negative thresholds denoted by red lines/horizontal lines in print). Linear regression (locally estimated scatterplot smoothing) including 90% confidence limits are indicated in shading. Maximum values shown were >10.0 and set to 10.0 for analysis. Date of positive RT-PCR for the COVID-19kp group, median (min, max): 2020 − 03 − 23 (2020 − 03 − 11, 2020 − 03 − 31). The same date was arbitrarily assumed for the COVID-19ip group (which also coincided with the first lockdown in Germany). Abbreviations. COVID-19ip, COVID-19 history unknown before program but identified as positive during the program; COVID-19kp, positive COVID-19 history known before program; COVID-19neg, COVID-19 history unknown before program and not identified during the program; IgG, immunoglobulin G; RT-PCR, reverse transcription polymerase chain reaction.
![Figure 4. IgG against S1 protein (Euroimmun) over time. Panel (A) Participants with COVID-19 history known before the program (n = 20), slope per day (confidence intervals): –0.08 (–0.12, –0.03); (B) Participants with COVID-19 history identified during the program (n = 10); (C) Comparison of levels in all participants (n = 141) over all four visits. *An optical density [OD] ratio of serum sample/OD of calibrator below 0.8 was evaluated as negative, 0.8–<1.1 as borderline and >1.1 as positive (positive/negative thresholds denoted by red lines/horizontal lines in print). Linear regression (locally estimated scatterplot smoothing) including 90% confidence limits are indicated in shading. Maximum values shown were >10.0 and set to 10.0 for analysis. Date of positive RT-PCR for the COVID-19kp group, median (min, max): 2020 − 03 − 23 (2020 − 03 − 11, 2020 − 03 − 31). The same date was arbitrarily assumed for the COVID-19ip group (which also coincided with the first lockdown in Germany). Abbreviations. COVID-19ip, COVID-19 history unknown before program but identified as positive during the program; COVID-19kp, positive COVID-19 history known before program; COVID-19neg, COVID-19 history unknown before program and not identified during the program; IgG, immunoglobulin G; RT-PCR, reverse transcription polymerase chain reaction.](/cms/asset/322c8521-fca6-4d47-8559-dde9812badbd/icmo_a_1914943_f0004_c.jpg)
In contrast to the Euroimmun ELISAs, the Diasorin CLIA, which detects IgG against both S1 and S2, showed a sustained IgG response over time () in the COVID-19kp group. In the COVID-19ip group (), the observed immune response against S1/S2 remained stable and persistent, although notably below the COVID-19kp group; however, the signal remained sufficiently large for a positive detection (). Positive and negative test results for IgG against S1/S2 proteins in participants over time are shown in Supplementary Table S10.
Figure 5. IgG against S1/S2 proteins (Diasorin CLIA) over time. Panel (A) Participants with COVID-19 history known before the program start (n = 20), slope per day (confidence intervals): 0.26 (–0.48, 1.00); (B) Participants with COVID-19 history unknown before the program but identified during the program (n = 10); (C) Comparison of levels in all participants (n = 141) over all four visits. *A level of <12 IU was considered negative; 12–<15 as borderline and ≥15 as positive (positive/negative thresholds denoted by red lines/horizontal lines in print). Linear regression (locally estimated scatterplot smoothing) including 90% confidence limits are indicated in shading. Date of positive RT-PCR for the COVID-19kp group, median (min, max): 2020 − 03 − 23 (2020 − 03 − 11, 2020 − 03 − 31). The same date was arbitrarily assumed for the COVID-19ip group (which also coincided with the first lockdown in Germany). Abbreviations. CLIA, chemiluminescence immunoassay; COVID-19ip, COVID-19 history unknown before program but identified as positive during the program; COVID-19kp, positive COVID-19 history known before program; COVID-19neg, COVID-19 history unknown before program and not identified during the program; IgG, immunoglobulin G; RT-PCR, reverse transcription polymerase chain reaction.
Individual graphs for participants over the study duration and long-term follow-up (>7 months; not described) are shown in Supplementary Figures S4 and S5.
Performance of diagnostic tests
Specificity, sensitivity, positive and negative predicted values are described in the Supplementary Appendix (Supplementary Table S11). Of note, since there were no new incidents of infection during the testing period, these findings should be interpreted accordingly.
BI HealthTracker App
Overall, 128 (90.8%) of the 141 participants used the BI HealthTracker App. As expected, due to the low incidence rate of COVID-19 in Germany during the testing period, only few symptoms potentially indicative for SARS-CoV-2 exposure were recorded by the App. These were followed up by the medical team and were found to be likely associated with hay fever, allergic asthma exacerbations and/or infections other than with SARS-CoV-2.
Discussion
No new infections of COVID-19 were detected during the pilot phase of COVID-19 Testing@BI, which was not unexpected given that the program started approximately 6 weeks after the first peak of the COVID-19 pandemic in Germany. At this point, Germany was in full lockdown, which included extensive social distancing and hygiene measures, thus limiting the opportunity of infection. The pilot phase ended on 10 June 2020, when SARS-CoV-2 infections were still at a comparatively low level [Citation1]. The four cases with positive RT-PCR results at 4–6 weeks were not necessarily still infectious: residual viral RNA in the throat has been reported many weeks after onset of illness [Citation8]. Based on the participants’ history, we concluded that RT-PCR likely detected viral debris rather than intact virus of the infectious stage. This emphasizes the variability in sampling and testing for viral RNA and the importance of a full differential diagnostic assessment instead of relying on one single test method and result.
In the two COVID-10kp participants who never developed any detectable antibodies, it is possible the SARS-CoV-2-specific antibody response waned or was never mounted in the first place. Lack of antibody response following positive RT-PCR has been reported before [Citation7], and it remains unclear whether immunity has been established in these cases. Importantly, both participants showed a prolonged and more severe course of disease with a longer period of convalescence.
It is plausible that participants medically judged to have had COVID-19 prior to testing belong to the particularly relevant group of COVID-19 “silent spreaders” who are more likely to transmit the infection since they do not feel sick and hence fail to isolate themselves. Indeed, up to 20% of people with SARS-CoV-2 infections have been reported to be asymptomatic [Citation9]. Importantly, the ability of LFAs and Euroimmun ELISAs to detect these participants differed markedly from detecting participants from the COVID-19kp group, in contrast with the Diasorin CLIA, which had a comparably high sensitivity for both cohorts. Given that the EuroImmun IgG assay (which measures S1 IgG) was mostly negative in these participants and that the Diasorin CLIA measures both S1 and S2 IgG, our findings suggest that COVID-19ip participants predominantly produced IgG antibodies recognizing S2 only. The S1 and S2 are part of the spike protein; thus, it is understandable that this group was not detected by the LFAs, which are directed against the N protein. Failure to detect recovered individuals with evident antibody levels against S1 and S2 protein of SARS-CoV-2 reveals an important limitation of antibody screening solely by LFA. In addition, not testing for S2 antibodies may lead to an under-reporting of cases. These observations had important implications for our strategy for the planned next-phase, large-scale testing of employees, and we concluded that LFAs for early antibodies were not suitable for our purpose.
Modulation of disease severity by antibodies against SARS-CoV-2, longevity of the antibody response and protection against re-infection are currently key areas of interest, in particular with the ongoing roll-out of vaccines [Citation2]. In our study, IgA and IgG levels measured by ELISA/CLIA were generally higher in COVID-19kp participants compared with COVID-19ip participants. This suggests that the latter group either produced lower levels of SARS-CoV-2 antibodies or that they were captured at a later stage in the course of infection, towards the end of the detectable period. Since COVID-19ip participants were assumed to have had milder disease, fewer symptoms may correlate with lower amounts of antibodies; indeed, a weaker immune response has been described in asymptomatic individuals [Citation7]. Fewer symptoms in participants may have reflected an exposure to a low viral load, with a corresponding weaker immune response [Citation10]. An alternative interpretation suggests that COVID-19ip participants may have had mild or asymptomatic SARS-CoV-2 infection with an earlier date of infection (e.g. January or early February 2020) and therefore missed the diagnostic window for Ig detection against the S1 protein during the observation period. Whilst we cannot say for certain and further studies are required, it raises the interesting possibility that the different antibody patterns observed between both groups may be driven by varying ability to produce antibodies against the S2 protein, with potential clinical consequences with regards to disease progression and recovery. On the other hand, it is entirely possible that the antibody response observed in our study may be independent of the S2 antigen, and instead, as mentioned above, may reflect a lower viral load at the point of infection, or differences in the immune function due to age or comorbidities.
In comparison with COVID-19kp participants, COVID-19ip participants had a higher median age. Other demographics and baseline characteristics were broadly similar between both groups. We found no difference in blood group type or cardiovascular disease history among any of our participant groups.
Our study has strengths and limitations. A strength of our study is that we identified three distinct groups (COVID-19kp, COVID-19ip and COVID-19neg) from whom we have collected comprehensive baseline demographics and longitudinal data on symptoms, viral RNA and IgA/M/G antibodies against different virus proteins. Moreover, our data can be regarded as real-world evidence. Limitations of our study are that there were no incident infections during the testing period due to lockdown and hygiene measures and that the data are based on a small, defined population (141 employees of BI in Germany); however, this population size was appropriate to define a testing strategy for the next (large-scale) testing phase. Further studies are required to uncover a potential mechanism for the differences in IgG formation that we report. Moreover, we cannot exclude that our observations may be due to higher sensitivity of the Diasorin S1/S2 IgG CLIA over the Euroimmun S1 IgG ELISA; however, given that both assays are accredited and commercially available, we believe that our findings are noteworthy.
Conclusion
This occupational healthcare program was designed to screen and monitor employees who have to work onsite for business continuity for essential medicine development and production. We found that LFAs for antibodies (IgM and IgG, which are currently available and commonly used for public health screening) may not always reliably detect current, recent or past infections; consequently, these have not been included in the upscaling of the program. An IgA LFA for detection of early antibody formation to judge current/recent infection with potential infectiousness could be relevant; however, this is not currently obtainable. Results from our pilot study suggest that regular targeted testing strategies using combinations of detection of viral RNA and, for example, in addition to PCRs with antigen-LFAs plus antibodies directed against different SARS-CoV-2 proteins, alongside a comprehensive baseline assessment of medical history and likelihood of infection, might be relevant for effective workforce screening and monitoring.
Transparency
Declaration of funding
Conduct of the research was funded by Boehringer Ingelheim as part of an occupational healthcare program performed by the occupational healthcare services.
Declaration of financial/other relationships
The authors declare no conflict of interest with regards to this work.
Peer reviewers on this manuscript have received an honorarium from CMRO for their review work but have no other relevant financial relationships to disclose.
Author contributions
PMZ, CK and EM were involved in the conceptional aspects of the study (planning and design). All authors contributed equally to the writing and critical review of the manuscript, including the first draft, provided final approval of the version to be published and agree to be accountable for all aspects of the work.
PMZ_suppl_clean_30Mar21.docx
Download MS Word (677.5 KB)Acknowledgements
The authors would like to thank all participants for taking part in the study.
The authors would like to thank the following contributors for their technical support and help with the study design and data analysis: BI: Timo Ahland, Susanne Allgäuer, Anna Arndts, Martin Beck, Ursula Bausenhart, Nicole Bauer, Gabriele Befurt, Claudia Blum, Benjamin Britz, Gudrun Buir, Julia Eggers, Carolyn Formella, Sybille Gondro, Goetz Gortner, Regina Gretzinger, Heike Haas, Andre Habierski, Hartmut Haeselbarth, Ulrike Hagel, André Haugg, Ulrike Hein-Rusinek, Marcel Irle, Tanja Isele, Carina Ittrich, Ines Jakob, Karin von Jeinsen, Maren Kaiser, Birte Kaupert, Sigrid Keller, Rainer Kirch, Miriam Klick, Kevin Knoche, Katja Koch, Liliana Kozic, Sascha Kreischer, Hildegard Kremper, Manuela Kreuder, Paul Kreye, Cornelia Kunz, Thorsten Lamla, Tobias Langbrugger, Berenike Lange, Rainer Lecht, Jan Launhardt, Martin Locher, Lorenz Maier, Show Reddy Maramreddy, Hanna-Marie Lustenberger, Ralf Minkenberg, Anja Mirau, Hagen Moerbel, Stefan Oelmeier, Liliane Olesch, Jennifer Otto, Erika Plenz, Alana Pottmeier, Barbara Raadatz, Sabrina Reimers, Maren Reinecke, Kerstin Saar, Saide Sahin, Eliane Sans, Antonie Sauter, Daniel Schwenk, Matthias Trampisch, Christina Vogel, Anke Webler-Messenger, Thomas Weissschuh, Maximilian Zahn, Sabrina Zech, Emil Zieglmaier, Christina Zimmermann, Sabine Zimmermann and Kai Zuckschwerdt; Bioscientia: Katrin Donde, Oliver Harzer, Uta Kuesters.
Medical writing support was provided by Kristina Standeven of MediTech Media, funded by Boehringer Ingelheim, under the guidance of the authors.
Data availability statement
None.
References
- Robert Koch-Institut. Robert Koch-Institut: COVID-19-Dashboard 2021 [cited 2021 Feb 28]. Available from: https://experience.arcgis.com/experience/478220a4c454480e823b17327b2bf1d4.
- Röltgen K, Powell AE, Wirz OF, et al. Defining the features and duration of antibody responses to SARS-CoV-2 infection associated with disease severity and outcome. Sci Immunol. 2020;5(54):eabe0240.
- Azkur AK, Akdis M, Azkur D, et al. Immune response to SARS-CoV-2 and mechanisms of immunopathological changes in COVID-19. Allergy. 2020;75(7):1564–1581.
- Arevalo-Rodriguez I, Buitrago-Garcia D, Simancas-Racines D, et al. False-negative results of initial RT-PCR assays for COVID-19: A systematic review. PLoS One. 2020;15(12):e0242958.
- Gudbjartsson DF, Norddahl GL, Melsted P, et al. Humoral immune response to SARS-CoV-2 in Iceland. N Engl J Med. 2020;383(18):1724–1734.
- Wajnberg A, Amanat F, Firpo A, et al. Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science. 2020;370(6521):1227–1230.
- Long QX, Tang XJ, Shi QL, et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med. 2020;26(8):1200–1204.
- Perera RAPM, Tso E, Tsang OTY, et al. SARS-CoV-2 virus culture and subgenomic RNA for respiratory specimens from patients with mild coronavirus disease. Emerg Infect Dis. 2020;26(11):2701–2704.
- Pollock AM, Lancaster J. Asymptomatic transmission of COVID-19. BMJ. 2020;371:m4851.
- Young MK, Kornmeier C, Carpenter RM, et al. IgG antibodies against SARS-CoV-2 correlate with days from symptom onset, viral load and IL-10. medRxiv. 2020. DOI:10.1101/2020.12.05.20244541.

