ABSTRACT
The Swedish Mountain (Fjäll) cattle and the subpopulation Fjällnära originates from northern Sweden. They are differentiated from both traditional and commercial cattle from southern Sweden. We analysed data from the GGP HD150k SNP array and investigated genetic diversity and differences between Fjäll and Fjällnära, and between different groups of Fjällnära. We found that the Fjällnära can be divided into four groups, which are differentiated from each other and from the other Fjäll cattle. We also compared allele frequencies between different groups of Fjäll and Fjällnära for some of the functional SNPs included in the SNP array. Interestingly, the B allele of Beta lactoglobulin has a very high frequency (94%) in the Fjällnära cattle, which is significantly different from the frequency in the Fjäll cattle (60%). Comparing older samples of Fjäll cattle with more recent samples, we found a significant increase in the frequency of the B allele in Fjäll cattle.
Introduction
The Swedish Mountain cattle (Fjäll) is a native breed with ancient history that once played an important role for milk production in northern Sweden. The first breeding centres were started in 1905 and bulls from the breeding centres were distributed to other parts of northern Sweden to improve the breed (Eriksson, Citation1943). In spite of its small current population size, the breed still displays a relatively high genetic diversity (Upadhyay et al., Citation2019). The Fjäll breed (also called Fjällko) is only distantly related to modern European breeds and has been shown to differ from the large commercial breeds in, e.g. frequency of milk casein alleles (Lien et al., Citation1999; Poulsen et al., Citation2017). The breed is said to be healthy and robust, with a relatively low milk production but a rather high milk fat and protein content. It is grouped together with other polled local breeds in the milk recording scheme, and together they show milk fat and protein contents similar to that of the Swedish Red, which is higher than for Swedish Holstein (Växa Sverige, Citation2019). Besides a large founder population, subdivision of the original population in local hard-to-reach mountain areas may have contributed to the conservation of genetic variation. Originally, the cattle in northern Sweden displayed a wide variety in colours, but with the formation of a breed standard in 1893, preference was given to polled Fjäll cattle of white coat colour with red or black spots.
The two polled local Swedish breeds Fjäll and Swedish Red polled (Rödkulla) were merged to form the Swedish Polled breed (Svensk Kullig Boskap, SKB) in 1938. Purebred animals of the original two breeds were still kept to a large extent, and since 1995 there is once more a separate Fjäll breed association. Some Swedish Polled animals with a large proportion of the Fjäll breed were eligible to enter the new herd book for Fjäll cattle. Genetic studies based on microsatellite markers have shown that the Swedish Fjäll and Swedish Red polled were not that closely related (Kantanen et al., Citation2000; Tapio et al., Citation2006), and this was confirmed in a study on SNP-markers by Upadhyay et al. (Citation2019).
By the end of the 1980s, a subpopulation of Fjäll cattle that was less intensively selected for milk production was recognized and called Fjällnära cattle. Kept in remote mountain areas, these animals had been milked only for consumption at the farm and not for commercial purposes, and many of these animals had not previously been registered in a herd book. The Fjällnära animals were found on farms in Klövsjö, Funäsdalen, Biellojaure and Lillhärjåbygget, which are now considered the four founder herds. Later also a few animals from other places have been included in the Fjällnära group. In 2008, an association (Föreningen Äldre Boskap) was formed with the aim to preserve the traditional Fjällnära cattle. There have been some differences in opinion between members of this association and of the older Fjäll cattle association (Svensk fjällrasavel) regarding breeding strategies and which animals to categorize as Fjällnära. Analyses of mtDNA have shown differences between Fjällnära cattle and other Fjäll cattle, in that some haplotypes occur in one of the groups but not in the other (Kantanen et al., Citation2009). In the study based on SNP array data, Upadhyay et al. (Citation2019) showed some population substructuring within the Fjällnära cattle, besides that the breeds of mountain cattle type (Fjäll, Fjällnära, and Bohus Polled) were differentiated from both local and commercial breeds in southern Sweden, with the exception of Swedish Polled which partly originate from Fjäll cattle.
In this study, our aim was to investigate the Fjäll and Fjällnära population structure further and also compare allele frequencies for some well-known functional SNPs in genes for which we had genotypes available from the Geneseek 150k SNP array. We focused on SNPs in genes related to milk production, content and processing properties; Beta casein, Beta lactoglobulin, FGF2, and some recessive disorders such as Mulefoot, BLAD, Citrullinemia, and DUMPS. Genes related to milk processing properties are interesting to study since Fjäll cattle has been shown to have better milk coagulation properties than Swedish red (Poulsen et al., Citation2017). Beta casein variants are important for yield and quality in cheese making, which are favourably influenced by the B-allele of the Beta casein gene (Schaar et al., Citation1985). Beta lactoglobulin is the major whey protein in cow milk, for which the B variant is associated with good properties for dairy products such as cheese, and the B allele of the Beta lactoglobulin gene is associated with larger cheese yield (Aleandri et al., Citation1990; Wedholm et al., Citation2006). Variants in the gene FGF2 have been shown to be associated with fat yield and percentage, somatic cell score and productive life (Wang et al., Citation2008). Mulefoot is a condition where the claws are not separated (isolated syndactoly) and has been shown to be caused by mutations in the LRP4 gene (Drögemüller et al., Citation2007). Bovine leukocyte adhesion deficiency (BLAD) is an autosomal recessive disorder where a causative mutation (an A to G substitution) at position 383 in the CD18 gene has been detected in Holstein (Shuster et al., Citation1992). Deficiency of uridine monophosphate synthase (DUMPS) is an autosomal recessive disorder that causes early embryonic mortality. DUMPS is caused by a mutation of a C to a T at codon 405 in exon 5 of the UMP synthase gene (Schwenger et al., Citation1994). Citrullinaemia is an autosomal recessive disorder where the calf is born without symptoms, but develops symptoms and then die early in life. The mutation is a substitution of a C to a T (Dennis et al., Citation1989). We also describe the origin of the Fjällnära group and present genetic differences between the founder farms Klövsjö, Funäsdalen, Biellojaure and Lillhärjåbygget and their relationship to the Fjäll cattle.
Material and methods
Sample selection and DNA extraction
We have used 150k genotypes from the Swedish native cattle breed Fjäll cattle and the subpopulation Fjällnära previously studied by Upadhyay et al. (Citation2019) together with additional genotypes using the same SNP array for 16 AI Fjäll bulls used in breeding in recent years. A description of the samples can be found in . The additional samples were from animals born 2000–2015 (), and because the samples investigated in Upadhyay et al. (Citation2019) were born 1976–1997, we were also able to investigate possible changes in the Fjäll breed between different time periods.
Table 1. Number of samples from Fjäll and Fjällnära (Fjäll is also divided into old and new samples) and heterozygosity and polymorphic loci. Ho is observed heterozygozity, He is expected heterozygosity.
For the analysis of SNPs known to be related to traits, we included 24 samples each from Swedish Red and from Swedish Holstein-Friesian from Upadhyay et al. (Citation2019) as a comparison. These samples were from animals born in the 1970s to the 1990s. They were thus contemporary to the old Fjäll and Fjällnära samples in the study. The Fjällnära samples used here were 15 of the samples collected in the 1990s and previously included in a Nordic collaboration (published for example in Kantanen et al., Citation2009, Tapio et al., Citation2006) and one additional bull born 1991 from which we had an old blood sample at our department. Using information from documents from sampling in the 1990s and information from the breed associations, we were able to categorize the 16 Fjällnära according to which of the four founder herds they belonged to. Four of the Fjällnära originated from Klövsjö, five from Funäsdalen, three from Biellojaure, and four from Lillhärjåbygget. Of the 23 old Fjäll samples that were not categorized as Fjällnära, 19 were included in the old Nordic project (for example Kantanen et al., Citation2000, Citation2009; Tapio et al., Citation2006), and four were other old samples stored at the department. The new Fjäll samples were from semen obtained from Viking Genetics.
SNP genotyping and quality control
For DNA extraction of the additional 16 samples that are not described in Upadhyay et al. (Citation2019) we used a QIAsymphony automated platform (Qiagen) following the manufacturer’s instructions. DNA was quantified and quality-controlled using a NanoDrop 8000 Spectrophotometer (ThermoFisher Scientific).
DNA samples were genotyped using the GeneSeek® Genomic Profiler High-Density Bovine 150 K (GGP HD150 K) array. Genotypes were called using the GenomeStudio® software (Illumina, San Diego, CA, USA). SNPs assigned to sex chromosomes and unassembled contigs were removed. In the quality control, thresholds were set to ≥0.95 call rate for both individuals and SNPs, and a minor allele frequency (MAF) ≥ 0.05. All quality filtering steps were carried out using PLINK 1.9 (Purcell et al., Citation2007). Hardy Weinberg equilibrium based filtering was not applied as we wanted to study substructures in the population that could partly have been caused by inbreeding and genetic drift. After the filtering 106,756 SNPs remained in the final dataset.
Genetic diversity within and among subpopulations
Average observed (Ho) and expected (He) heterozygosity were estimated using the package adegenet (Jombart, Citation2008) in R (R Core Team, Citation2013). For principal component analysis (PCA), we used the ‘snpgdsPCA’ function of the SNPrelate package (Zheng et al., Citation2012, Citation2017). The proportion of polymorphic markers was calculated using PLINK 1.9 (Purcell et al., Citation2007). Note that for the unbiased estimation, the proportion of polymorphic markers was calculated before the filtering based on call rate and MAF, but SNPs located on sex chromosomes, mt, and unassembled contigs were removed.
To estimate genetic distance (Fst), ‘stamppFst’ function as implemented in an R package ‘StAMPP’ (Pembleton et al., Citation2013) was used. Later, ‘NJ’ function implemented in ‘phangorn’ R package was used to contruct a Neighbor-joining tree from the Fst values (Schliep, Citation2011). ADMIXTURE analysis was carried out with K values from 2 to 6 (Alexander et al., Citation2009). The python package PONG was used to visualize the ADMIXTURE result (Behr et al., Citation2016).
Comparison of allele frequencies for trait-associated SNPs
Allele frequencies for seven different SNPs previously shown to be related to milk quality or genetic defects in cattle were compared between the different subgroups of Fjäll cattle, and with Swedish Holstein-Friesian and Swedish Red.
The SNP called BCNAB identifies the B allele for Beta casein. The three SNPs BetaLact, BetaLact2 and BetaLactB1 identifies the B allele for the Beta lactoglobulin gene. The Fjäll breed organization had already genotyped some of the AI bulls for these two genes (Svensk Fjällrasavel, Citation2019), and in this study we provide SNP genotypes of additional animals. Because the gene variant carried by 19 of the included bulls were known prior to this study, we could easily conclude which SNP alleles correspond to the which known alleles of these genes in the Fjäll and Fjällnära cattle. The B allele of Beta casein is allele G at BCNAB in the forward allele and in the top allele. The Beta lactoglobulin B allele is the C allele in the forward allele and G in the top allele of all three SNP assays.
For the SNP (SNP name Mulefoot-241) in the gene LRP4 associated with mulefoot (Drögemüller et al., Citation2007) it is the A allele at this SNP that is causing mulefoot if homozygous. The SNP name on the 150k SNP array for Bovine leukocyte adhesion deficiency is BLAD, that for citrullinemia is Citrullinemia_3 and that for Deficiency of uridine monophosphate synthase is DUMPS. The descriptions of the SNPs associated with traits can be found on Neogen’s website (https://genomics.neogen.com/pdf/slicks/ggp_bovine150k.pdf). To test if the genotype distributions were significantly different in different groups of animals, a Fisheŕs exact test was used, with the command fisher.test in R (R Core Team, Citation2013).
Results and discussion
Measures of genetic diversity
The proportion of polymorphic loci, observed heterozygosity (Ho) and expected heterozygosity (He) were higher in Fjäll than in Fjällnära cattle (). The Ho was smaller than He in Fjällnära (0.3145 vs 0.3727), indicating more inbreeding than in Fjäll where Ho was slightly higher than He in both old and new samples of Fjäll. This could be expected, as the Fjäll population is considerably larger than the Fjällnära, which was re-created from animals from a few farms. Thus, genetic drift is likely to have played larger role in lowering the genetic diversity in Fjällnära. The inbreeding (FIS) in the populations calculated as (He−Ho)/He was 0.156 in Fjällnära and −0.024 in Fjäll. Since the Fjällnära herds were small, matings of relatives have certainly taken place and another factor contributing to the high FIS is the population structure within the Fjällnära that is described in the next section. In this study where we have added 16 additional more recent samples from Fjäll individuals compared to the study of Upadhyay et al. (Citation2019), we could compare the old and new samples of Fjäll. The newer samples (born 2000–2015) had a bit lower values of both observed and expected heterozygosity compared to the old samples (born 1976–1997). Note however that the newer samples are all bulls whereas the old samples were mostly from cows. As the selection intensity for bulls is higher than for cows, the males would be expected to be a bit ahead of the females and therefore, the rate of change in heterozygosity per time unit may be somewhat biased. The breed organization is actively striving to avoid matings of close relatives, and to conserve genetic variation in the breed, and it is a good sign that the Fjäll cattle (excluding the Fjällnära) has no signs of inbreeding and has not lost so much genetic diversity over the last decades according to our results.
Substructures in the Fjäll and Fjällnära cattle
The PCA plot () shows that the old and the new Fjäll breed samples overlap. The Fjällnära samples are subdivided into different clusters that correspond to the farms of origin. The first principal component clearly separates the three Biellojaure samples from the other Fjällnära and from the Fjäll individuals. The second principal component clearly separate the four Lillhärjåbygget samples in the bottom, four of the five Funäsdalen samples and the four Klövsjö samples in the middle and the old and the new Fjäll samples together with one Fjällnära from Funäsdalen in the top. Even though the Klövsjö samples and four of the Funäsdalen individuals cluster together, they are not totally intermixed and the four Klövsjö samples are lower in the second principal component than the Funäsdalen samples (). We found out from studying pedigree data and a document written at the time of sampling that the Funäsdalen cow that clustered together with the Fjäll samples had a Fjäll AI bull as a sire, whereas the other four were from Fjällnära sires (not identified) born in their own farm. Traditionally, owners of Fjällnära animals often kept several bulls and did not control which bulls were the sire of which calf. The first component explains 7.62% of the variation, the second component explains 5.81% and the third component explain 4.65% of the variation.
Figure 1. PCA analysis of Fjäll and Fjällnära. The Fjäll samples are divided into the old (SMC_O) samples from animals born 1976-1997 and new (SMC_N) samples from animals born 2000-2015. The Fjällnära samples are divided into the founder farms they originate from. FNC_B (upper right corner) is from Biellojaure, FNC_F (around PC1 0.03, PC2 -0.1) is from Funäsdalen, FNC_K (around PC1 0.05, PC2 -0.15) is from Klövsjö, and FNC_L (around PC2 -0.4) is from Lillhärjåbygget.
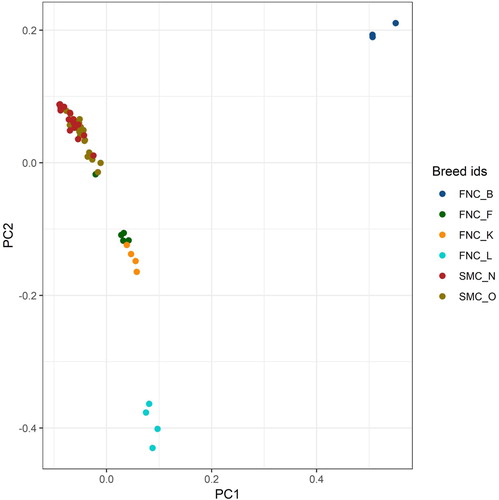
FST values between the different groups of Fjällnära and between Fjällnära groups and Fjäll were high and significant. The FST values between the groups can be seen in . The highest FST (0.395) was between Fjällnära from Biellojaure and Fjällnära from Lillhärjåbygget. In agreement with the PCA results in the Fjällnära group from Funäsdalen had lower FST to old and newer samples from Fjäll cattle (0.072 and 0.083 respectively) than the other Fjällnära groups. The FST between the older and newer samples of Fjäll cattle was very low (0.008).
Table 2. FST values between groups of Fjällnara and old and new Fjäll.
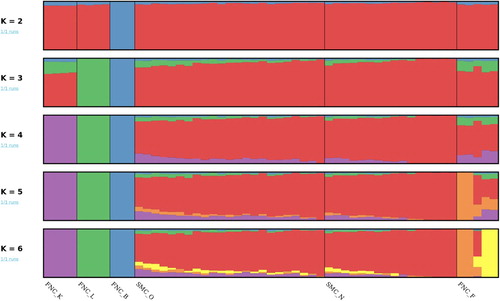
The results from the ADMIXTURE analysis show the same pattern of population structure as the PCA analysis and the FST, with the Fjällnära group Biellojaure separating from the others at K = 2 and older and newer samples of Fjäll being similar to each other at all tested values of K (). Fjällnära from Lillhärjåbygget forms it’s own group from K = 3, and Klövsjö from K = 4. The Funäsdalen cow with a Fjäll AI bull as a sire that clustered together with Fjäll in the PCA plot is also in the ADMIXTURE analysis showing a mixed ancestry (most clearly seen at K = 6).
Figure 2. The ADMIXTURE result with K ranging from 2 to 6. The Fjäll samples are divided into the old (SMC_O) samples from animals born 1976–1997 and new (SMC_N,) samples from animals born 2000–2015. The Fjällnära samples are divided into the founder farms they originate from. FNC_B is from Biellojaure, FNC_F is from Funäsdalen, FNC_K is from Klövsjö, and FNC_L is from Lillhärjåbygget.
Also in the neighbour joining tree the older and newer samples of Fjäll are very similar to each other and all the four Fjällnära group show long branch lengths indicating differentiation of the Fjällnära groups (Supplementary Figure 1). The differentiation of the Fjällnära groups is probably mostly due to genetic drift due to small populations being isolated.
Differences in allele frequencies of trait-associated SNPs
We investigated differences in allele frequencies for a few functional SNPs in the Fjäll and Fjällnära cattle breeds and as a comparison in Swedish red and Swedish Holstein Friesian (). Our results show that the B allele of the Beta casein gene is rare among the Swedish mountain cattle breeds. It was only found in four of the investigated younger (born 2000 and after) AI bulls of the Fjäll breed. This corresponds to an allele frequency of 5% among the total Fjäll samples and 12.5% among the new Fjäll samples. This is in agreement with the 13% frequency of the B variant of the protein in the milk of the Fjäll breed in the study of Poulsen et al. (Citation2017). The B allele was not found in any of the older samples from Fjäll (although one had missing value for this SNP) and not in any of the samples of Fjällnära (two of the samples had missing values), Swedish Red or Swedish Holstein-Friesian. In the study of Gustavsson et al. (Citation2014), the allele frequency was 0.9% among Swedish Red and 4.6% among Danish Holstein.
Table 3. Allele frequencies of SNPs related to functional traits and within parentheses the number of animals being homozygous or heterozygous for the allele.
The B allele in Beta lactoglobulin on the other hand, was much more common in our data. It was found at an allele frequency of 60% in Fjäll, and 94% in Fjällnära individuals in the study. The sampled Fjällnära individuals originating from the farms in Biellojaure, Lillhärjåbygget and Klövsjö were all homozygous for the B allele. Among the individuals from Funäsdalen, two were heterozygous and three were homozygous for the B allele. It is notably that the B allele was not fixed in the Funäsdalen group among Fjällnära, and it agrees with the finding that Funäsdalen is the Fjällnära group closest to Fjäll in the PCA (). The difference in frequency between Fjäll and Fjällnära was significant (p = 0.0016).
The Beta lactoglobulin B allele was more common among the Fjäll bulls born 2000–2015 (72%) than among the older Fjäll samples (52%). This difference between old and more recent Fjäll breed samples was also significant (p = 0.028). The frequency of the B allele for Beta lactoglobulin has thus increased over the last decades. This is not surprising as the breed association has genotyped bulls and selected for the B allele because of its favourable effects on the milk properties (Svensk Fjällrasavel, Citation2019). The breed association Svensk Fjällrasavel wants to emphasize the breed’s superiority for milk quality aspects. The frequency 72% among the more recent Fjäll samples is in agreement with the study of Poulsen et al. (Citation2017), which studied proteins in milk from 23 cows from this breed and found a frequency of the B variant of 71.7%. The frequencies in Swedish Red and Swedish Holstein-Friesian were 52% and 46%, respectively, in this study. It should be noted that the individuals from these breeds studied were from the 1970s to 1990s and, frequencies could be different in now living animals of these breeds.
It is interesting that the B allele of Beta lactoglobulin had such high frequency in Fjällnära. A possible explanation for this is that these farms were located far away from necessary infrastructure to collect milk for dairies, and therefore all milk and derived products were consumed locally. It was therefore not seen as an advantage if the cow produced large amounts of milk, but instead, it was preferred to produce milk suitable for making storable products such as cheese. The breeding strategies among these farmers were thus probably different from those at farms delivering milk to dairies that received larger payment for more milk.
In the FGF2 SNP, there was no significant difference between Fjäll and Fjällnära. The SNP allele A had a frequency of 42% in Fjäll and 34% in Fjällnära. The difference between old and new Fjäll for this allele was also not significant (37% and 50%, respectively). In the Swedish Red, the frequency was 31% and Swedish Holstein-Friesian had a frequency of 61%.
Another very interesting gene for milk properties is CSN3 that codes for Kappa casein that is important for cheese production (Wedholm et al., Citation2006). Poulsen et al. (Citation2017) showed that the B variant of kappa casein is the most common in Fjäll cattle whereas the A variant is the most common in Swedish Red Polled. Unfortunately, we did not have genotypes for this gene in our dataset. For future studies, it would be very interesting to look more in detail at all the casein variants to study possible differences between the subpopulations of Fjäll cattle.
It is since long known that the Fjäll breed had problems with gonadal hypoplasia (Eriksson, Citation1943; Settergren, Citation1964). A recent study identified the causative mutation in Fjäll and in Northern Finncattle (Venhoranta et al., Citation2013). For the four other defects we investigated in this paper we found a defect allele in Fjäll cattle in only one individual. The mulefoot allele was found in one heterozygote of the Fjäll breed (a female born 1991). It was not found in any of the Fjällnära or Swedish Holstein-Friesian, but interestingly it was quite common in these older samples from Swedish Red (19%), collected before genotyping of breeding animals became common practise. The defect alleles for BLAD, Citrullinemia and DUMPS were not found in any of the samples of Fjäll, Fjällnära or Swedish Red. One old Swedish Holstein-Friesian cow was heterozygotic for the BLAD allele.
Conclusion
The frequency of the B allele for the Beta lactoglobulin gene, of importance for cheese production, seems to have increased in the Swedish native Fjäll cattle as a result of selection bases on genotype information. The general amount of detected genetic diversity was similar in more recent and older samples from this breed, however, and the analyses of population structure showed that they are very similar. The Fjäll and Fjällnära cattle are genetically differentiated, and the Fjällnära show a significantly higher allele frequency of the B allele for the Beta lactoglobulin gene, close to fixation. Within the Fjällnära group, genetic differentiation corresponds to different locations of origin, where samples belonging to two of the founding farms were more closely related than the other. For the few studied markers for genetic defects, the frequencies of unfavourable alleles were very low in the Fjäll and Fjällnära samples, which is in agreement with reputation of these populations as being healthy. Other defects may segregate in these populations however, which were not included in this study.
Acknowledgements
We gratefully acknowledge contributions of samples and information about breeds and animals from the different breed organizations, individual animal owners, and VikingGenetics. We also acknowledge Kaj Sandberg and Birgitta Danell for coordinating and building a sample collection of some of the old samples in earlier projects. We want to thank Susanne Gustafsson, Gabriela Bottani Claros, and the SLU Biobank for help with DNA extraction and concentration measure.
Disclosure statement
No potential conflict of interest was reported by the authors.
ORCID
Anna M. Johansson http://orcid.org/0000-0002-9762-0497
Susanne Eriksson http://orcid.org/0000-0003-3357-5065
Additional information
Funding
References
- Aleandri, R., Buitazzoni, L.G. & Schneider, J.C. (1990). The effects of milk protein polymorphisms on milk components and cheese-producing ability. J. Dairy Sci. 73, 241–255. doi: 10.3168/jds.S0022-0302(90)78667-5
- Alexander, D.H., Novembre, J. & Lange, K. (2009). Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664. doi:10.1101/gr.094052.109.
- Behr, A.A., Liu, K.Z., Liu-Fang, G., Nakka, P. & Ramachandran, S. (2016). Pong: Fast analysis and visualization of latent clusters in population genetic data. Bioinformatics 32, 2817–2823. doi:10.1093/bioinformatics/btw327.
- Dennis, J.A., Healy, P.J., Beaudet, A.L. & O'Brien W.E. (1989). Molecular definition of bovine argininosuccinate synthetase deficiency. Proc. Natl. Acad. Sci. USA 86, 7947–7951. doi: 10.1073/pnas.86.20.7947
- Drögemüller, C., Leeb, T., Harzelius, B., Tammen, I., Distl, O., Höltershinken, M., Gentile, A., Duchesne, A. & Eggen, A. (2007). Congenital syndactyly in cattle: Four novel mutations in the low density lipoprotein receptor-related protein 4 gene (LRP4). BMC Genetics 8, 5. doi:10.1186/1471-2156-8-5.
- Eriksson, K. (1943). Hereditary Forms of Sterility in Cattle. Biological and Genetical Investigations. (Lund: Håkan Ohlssons Boktryckeri), p. 155.
- Gustavsson, F., Buitenhuis, A.J., Johansson, M., Bertelsen, H.P., Glantz, M., Poulsen, N.A., Lindmark-Månsson, H., Stålhammar, H., Larsen, L. B., Bendixen, C., Paulsson, M. & Andrén, A. (2014). Effects of breed and casein genetic variants on protein profile in milk from Swedish Red, Danish Holstein, and Danish Jersey cows. J. Dairy Sci. 97, 3866–3877. doi: 10.3168/jds.2013-7312
- Jombart, T. (2008). Adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics. 24, 1403–1405. doi: 10.1093/bioinformatics/btn129
- Kantanen, J., Olsaker, I., Holm, L. E., Lien, S., Vilkki, J., Brusgaard, K., Eythorsdottir, E., Danell, B. & Adalsteinsson, S. (2000). Genetic diversity and population structure f 20 north European cattle breeds. J. Hered. 91 (6), 446–457. doi: 10.1093/jhered/91.6.446
- Kantanen, J., Edwards, C.J., Bradley, D.G., Viinalass, H., Thessler, S., Ivanova, Z., Kiselyova, T., Cinkulov, M., Popov, R., Stojanovic, S., Ammosov, I. & Vilkki, J. (2009). Maternal and paternal genealogy of Eurasian taurine cattle (Bos Taurus). Heredity 103, 404–415. doi: 10.1038/hdy.2009.68
- Lien, S., Kantanen, J., Olsaker, I., Holm, L.-E., Eythorsdottir, E., Sandberg, K., Dalsgard, B. & Adalsteinsson, S. (1999). Comparison of milk protein allele frequencies in Nordic cattle breeds. Anim. Genet. 30, 85–91. doi: 10.1046/j.1365-2052.1999.00434.x
- Pembleton, L.W., Cogan, N.O.I. & Forster, J.W. (2013) StAMPP: An R package for calculation of genetic differentiation and structure of mixed-ploidy level populations. Mol. Ecol. Resour. 13, 946–952. doi:10.1111/1755-0998.12129.
- Poulsen, N.A., Glantz, M., Rosengaard, A.K., Paulsson, M. & Larsen, L.B. (2017). Comparison of milk protein composition and rennet coagulation properties in native Swedish dairy cow breeds and high-yielding Swedish Red cows. J. Dairy Sci. 100, 8722–8734. doi: 10.3168/jds.2017-12920
- Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M.A.R., Bender, D., Maller, J., Sklar, P., de Bakker, P.I., Daly, M.J. & Sham, P.C. (2007). PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hu. Gene. 81, 559–575. doi: 10.1086/519795
- R Core Team (2013). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing (Vienna, Austria). Available at: http://www.R-project.org/.
- Schaar, J., Hansson, B. & Pettersson, H.E. (1985). Effects of genetic variants of κ-casein and β-lactoglobulin on cheesemaking. J. Dairy Res. 52, 429–437. doi: 10.1017/S002202990002433X
- Schliep K.P. (2011). Phangorn: Phylogenetic analysis in R. Bioinformatics 27, 592–593. doi:10.1093/bioinformatics/btq706.
- Schwenger, B., Tammen, I. & Aurich, C. (1994). Detection of homozygous recessive genotype for deficiency of uridine monophosphate synthase by DNA typing among bovine embryos produced in vitro. J. Reprod. Fertil. 100, 511–514. doi: 10.1530/jrf.0.1000511
- Settergren, I. (1964). The Ovarian Morphology in Clinical Bovine Gonadal Hypoplasia with Some Aspects of its Endogrine Relations (Stockholm: Stockh Almqvist Wiksells), p. 108.
- Shuster, D.E., Kehrli, M.E. Jr., Ackermann, M.R. & Gilbert, R.O. (1992). Identification and prevalence of a genetic defect that causes leukocyte adhesion deficiency in Holstein cattle. Proc. Natl. Acad. Sci. U.S.A. 89, 9225–9229.
- Svensk Fjällrasavel. (2019). Available at September 2019 http://www.fjallko.se/mjolkkaseiner/ostkaseiner
- Tapio, I., Värv, S., Bennewitz, J., Maleviciute, J., Fimland, E., Grislis, Z., Meuwissen, T.H.E., Miceikiene, I., Olsaker, I., Viinalass, H., Vilkki, J. & Kantanen, J. (2006). Prioritization for conservation of northern European cattle breeds based on analysis of microsatellite data. Conserv. Biol. 20, 1768–1779. doi: 10.1111/j.1523-1739.2006.00488.x
- Upadhyay, M., Eriksson, S., Mikko, S., Strandberg, E., Stålhammar, H., Groenen, M.A.M., Crooijmans, R.P.M.A., Andersson, G. & Johansson, A.M. (2019). Genomic relatedness and diversity of Swedish native cattle breeds. Genet. Sel. Evol. 51, 56. doi: 10.1186/s12711-019-0496-0
- Växa Sverige. (2019). Cattle Statistics 2019. 39. Available at September 2019 https://www.vxa.se/globalassets/dokument/statistik/husdjursstatistik-2019.pdf
- Venhoranta, H., Pausch, H., Wysocki, M., Szczerbal, I., Hänninen, R., Taponen, J., Uimari, P., Flisikowski, K., Lohi, H., Fries, R., Switonski, M. & Andersson, M. (2013). Ectopic KIT copy number variation underlies impaired migration of primordial germ cells associated with gonadal hypoplasia in cattle (Bos Taurus). PLoS ONE 8(9), e75659. doi: 10.1371/journal.pone.0075659
- Wang, X., Maltecca, C., Tal-Stein, R., Lipkin, E. & Khatib, H. (2008). Association of bovine fibroblast growth factor 2 (FGF2) gene with milk fat and productive life: an example of the ability of the candidate pathway strategy to identify quantitative trait genes. J. Dairy Sci. 91(6), 2475–2480. doi: 10.3168/jds.2007-0877
- Wedholm, A., Larsen, L.B., Lindmark Månsson, H., Karlsson, A.H. & Andrén, A. (2006). Effect of protein composition on the cheese-making properties of milk from individual dairy cows. J. Dairy Sci. 89, 3296–3305. doi: 10.3168/jds.S0022-0302(06)72366-9
- Zheng, X., Levine, D., Shen, J., Gogarten, S.M., Laurie, C. & Weir, B.S. (2012).A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28, 3326–3328. doi: 10.1093/bioinformatics/bts606
- Zheng, X., Gogarten, S.M., Lawrence, M., Stilp, A., Conomos, M.P., Weir, B.S., eLaurie, C. & Levine, D. (2017). Seqarray – a storage-efficient high-performance data format for WGS variant calls. Bioinformatics 33, 2251–2257. doi: 10.1093/bioinformatics/btx145

