ABSTRACT
A simple intracellular proteomic study was conducted to investigate the biological activities of Aspergillus niger during industrial enzyme production. A strain actively secreting a heterologous enzyme was compared to a reference strain. In total, 1824 spots on 2-D gels were analyzed using MALDI-TOF MS, yielding 343 proteins. The elevated levels of UPR components, BipA, PDI, and calnexin, and proteins related to ERAD and ROS reduction, were observed in the enzyme-producer. The results suggest the occurrence of these responses in the enzyme-producers. Major glycolytic enzymes, Fba1, EnoA, and GpdA, were abundant but at a reduced level relative to the reference, indicating a potential repression of the glycolytic pathway. Interestingly, it was observed that a portion of over-expressed heterologous enzyme accumulated inside the cells and digested during fermentation, suggesting the secretion capacity of the strain was not enough for completing secretion. Newly identified conserved-proteins, likely in signal transduction, and other proteins were also investigated.
Abbreviations: 2-D: two-dimensional; UPR: unfolded protein response; ER: endoplasmic reticulum; ERAD: ER-associated protein degradation; PDI: protein disulfide-isomerase; ROS: reactive oxygen species; RESS: Repression under Secretion Stress; CSAP: Conserved Small Abundant Protein; TCTP: translationally controlled tumor protein.
Graphical abstract
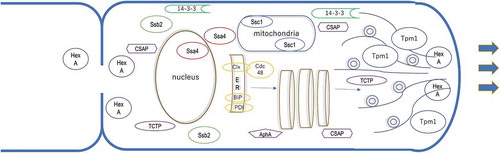
Detected major cell components in this study are illustrated in the predicted location. Thine allows show the secretion pathway. Thick allows indicate apical mycelial growth.
Aspergillus niger is an important industrial fungus used in the manufacturing of organic acids, such as citric acid, and industrial enzymes [Citation1–Citation3]. A. niger secretes several enzymes that decompose plant biomass, such as amylolytic, proteolytic (hemi) cellulolytic, pectinolytic enzymes, and oxidoreductases (catalase and glucose oxidase) [Citation4,Citation5]. Many of these enzymes are used safely in food processing [Citation1,Citation2]. These products and A. niger are recognized as “generally regarded as safe (GRAS)” by the United States Food and Drug Administration (FDA) [Citation1]. Their safety and ability to secrete enzymes in large amounts have rendered A. niger as popular host organisms for heterologous protein production [Citation6–Citation9]. Numerous enzymes and mammalian proteins, including bovine prochymosin and human interleukin-6 are expressed in A. niger [Citation7,Citation8]. A. niger can secrete native enzyme at high levels (>20 g/L) [Citation10], but yields of heterologous proteins are typically low [Citation7,Citation10]. Approaches that have been taken to address this problem of low yield [Citation7–Citation9] include over-expression of chaperones or foldases [Citation11], eliminating a protein degradation pathway [Citation12], and altering morphology [Citation13]. A recent trend has been to explore cellular activities for systematic control of protein production [Citation10,Citation14–Citation17].
The advancement and expansion of bioinformatics to embrace the new “omics,” namely; genomics, proteomics, transcriptomics, and metabolomics, has facilitated the comprehensive analysis of cellular processes and a better understanding of their associated cellular activities [Citation15–Citation18]. Because of their commercial importance, the genome sequence of two strains of A. niger has been determined: strain CBS513.88, the ancestor of the currently used enzyme-production strains [Citation19], and strain ATCC1015, known for citric acid production [Citation3]. This genomic information has led to a comprehensive omic analysis of this organism, including several transcriptomic analyses and proteomic studies [Citation10,Citation20–Citation27]. These studies have contributed to the understanding of certain cellular activities of A. niger strains producing enzymes, such as cellular responses to secretion stress [Citation10,Citation20,Citation25] or metabolic responses under oxygen-limited conditions [Citation26]. However, Aspergillus genes are far from fully characterized and the functions of many remain unknown. Furthermore, cellular activities that comprise a response to enzyme production and secretion will be many and complex, hence a significant amount of further investigation is required to achieve a full understanding of them.
Here we report a proteomic study of A. niger strains used for the production of industrial enzymes, in order to add to the information database and contribute to an understanding of the cellular activities involved in enzyme production. The study identifies several proteins at elevated expression levels during enzyme production, most of which are established components of a stress response. However, two uncharacterized abundant proteins were also identified, which seem to be conserved in Aspergillus and likely participate in signal transduction.
Materials and methods
Strains and samples for the analysis
Two strains of A. niger were used in this study. C1100 is a transformant that produces a heterologous glucoamylase from Trametes cingulate described in US8148127 [Citation28]. The strain was generated using an in-house A. niger strain derived from NN049184, which was originally isolated in Denmark from a soil sample. The endogenous glucoamylase gene was inactivated in C1100. The plasmid used for transformation comprised the cDNA clone of T. cingulate glucoamylase and an expression cassette. The cassette comprises the neutral amylase II promoter from A. niger fused to the 5ʹ-untranslated region (5ʹ-UTR) of triosephosphate isomerase gene from A. nidulans, the glucoamylase terminator from A. niger and the selective marker gene, amdS, from A. nidulans. The second strain, C1844, was derived from NN059095 [Citation29] of NN049184 lineage. The endogenous glucoamylase gene and alpha-amylase genes were inactivated by gene disruption in C1844. Fermentations were conducted in lab-scale 5 L tanks (ABLE corporation, Tokyo) with fed-batch fermentation to mimic commercial enzyme production conditions [Citation30]. Culture broths were collected at 48, 72 and 120 h for preparation of the cell extract.
Preparation of cell extract
Cells were harvested from the culture broths by filtration through a paper filter, followed by immediately rinsing with plenty of water and dehydrated on a paper towel. The harvested mycelia were frozen in liquid nitrogen and kept at −80°C until analysis. Frozen mycelia were ground to powder under liquid nitrogen using a pre-chilled mortar. Ground cells were suspended in lysis buffer consisting of; urea 5.1 g, 10% (w/v) SDS 0.2 mL, 20% (v/v) Triton X-100 1.0 mL, DTT 0.1 g, Pharmalyte [Citation3–Citation10] (GE Healthcare Japan) 0.2 mL, and one tablet of cOmpleteTM Mini protease inhibitor cocktail (Roche product, Sigma-Aldrich/Merck, Japan), made up to 10 mL in Milli-Q water. The homogenate was incubated at 16°C for 20 min, and then centrifuged at 3500 rpm to collect the supernatant solution. Disulfide-bonds in proteins were reduced and alkylated using the ProteoPrep Reduction and Alkylation Kit (PROTRA, Sigma-Aldrich/Merck, Japan) according to the manufacturer’s protocol. Protein solutions were concentrated and purified using a ReadyPrepTM 2-D Cleanup Kit (BIO-RAD Laboratories, Inc., Japan). This process was repeated twice to obtain protein concentrations around 20 mg/mL. A 2-D Quant Kit (GE Healthcare Japan) was used for the determination of protein concentration in crude and purified protein solutions.
Two-dimensional gel electrophoresis
Two-dimensional (2-D) gel electrophoresis was conducted on two scales. A preliminary run was made on a small scale using the DiskRun system (Atto Corporation, Tokyo). A precast agarose gel column, agarGEL A-M310 (2.5 mm diameter x 75 mm, pH 3–10, Atto), was used for the first dimension of isoelectric focusing, and precast poly-acrylamide gel, e-PAGEL (12.5%T, 90 mm (W) × 83 mm (H) × 1 mm (T), Atto) was used for the second dimension of SDS-PAGE. Large-scale electrophoresis was conducted for better separation yielding more spots. The apparatus, CoolPhoreStar IPG-IEF (Anatech Co., Ltd., Tokyo) was used for immobilized pH-gradient isoelectric focusing in the first dimension and CoolPhoreStar SDS-PAGE Dual-200 K was used for SDS-PAGE in the second dimension. Electrophoresis was conducted following the procedure previously described by T. Toda accessed at: http://proteome.tmig.or.jp/2D/2DE_method.html. The separation was achieved by employing Immobiline® DryStrip pH3-10NL or DryStrip pH 4–7, 18 cm (GE Healthcare Japan) for the first dimension, and a manually made poly-acrylamide gel (12.5% T, 190 mm (W) x 175 mm (H) x 1 mm (T)) for the second-dimensional electrophoresis. After electrophoresis, gels were stained with Coomassie brilliant blue (CBB) for visualization of the protein spots. Images of 2-D gels were scanned by Xcise (Shimadzu Corporation, Kyoto). Spot intensity was analyzed using Image-J (NIH, https://imagej.nih.gov/ij/).
Mass Spectrometry and protein identification
Protein spots were machine-excised, destained, and digested with trypsin using Xcise (Shimadzu Corporation, Kyoto). The digested peptides were purified and concentrated with ZipTip® (Millipore/Merck, Japan) and extracted with 0.1% TFA-acetonitrile solution containing α-cyano-4-hydroxycinnamic acid, and finally spotted on to metal plates for mass analysis. The mass spectrometric analysis was carried out using a MALDI-TOF_MS AXIMA CFR Plus (Shimadzu. Kyoto). The peptide mass fingerprint was analyzed using the MASCOT Sever and the NCBInr database (updated in August 2016). Protein identification was described using the locus tag of A. niger CBS 513.88 [Citation19]. Information regarding predicted protein function and yeast homolog was sourced from AspDB (http://www.aspergillusgenome.org/) (up-dated in 2015).
Results and discussion
Proteomic analysis and protein detection
A proteomic study was made using two recombinant A. niger strains; C1100, the test strain which over-secretes a heterologous glucoamylase, and C1844, a reference strain, which is not producing a heterologous enzyme and whose major secreted protein genes have been deleted. Both strains were fermented in lab-scale tanks under fed-batch conditions, mimicking industrial enzyme productions. Culture samples at different time points, 48, 72 and 120 h, were collected and preparation of cell extract was attempted for two-dimensional (2-D) gel electrophoresis. However, the quality of the samples obtained at the later stage of fermentation was poor due to partial cell lysis; therefore, the 48-h samples were mainly used for the analysis. In total, 14 gels were made, and 1832 stained spots were analyzed by MALDI-TOF_MS. Of these, 1445 spots were successfully identified based on the obtained amino acid sequences, resulting in the detection of 343 proteins. Detected proteins were categorized based on their predicted or known functions (see for a summary). As most proteins in A. niger have not been characterized, many protein functions were predicted based on their similarity to or orthologues in other organisms. A full list of detected proteins in this study is shown in Table S1.
Table 1. Functional category of the identified proteins.
Comparison of proteins detected in the two strains
Visual comparison with differential 2-D gels
A comparison of protein profiles between two strains, an enzyme-producer and the reference strain, can identify proteins and associated cellular activities that are affected on enzyme production in A. niger. A better understanding of the effect of heterologous protein production on the organism could lead to strategies to improve the yield and is also of basic interest with respect to gaining molecular insight into underlying cellular response mechanisms.
A qualitative visual comparison was made by analysis of differential 2-D gels (pH4-7) with 48-h samples for each strain. A different color was used to visualize the spots and gel images for each, then overlaid for spot comparisons (). When spot intensity was stronger in one strain versus the other, the spot appears in a specific color. If spot intensity is equal in the two strains, the spot appears black. The image shown in displays a couple of spots in different colors, indicating that the concentrations of these proteins were significantly different between the two strains. The large green spots labeled HP in are the heterologous glucoamylase expressed in C1100. Other spots shown in green include BipA (An11g04180), calnexin (ClxA) (An01g08420) and protein disulfide isomerase (PDI) (An02g14800). They are chaperones and foldases assisting protein folding in the ER. Pink spots represent proteins produced at a higher level in C1844 and are identified as fructose bisphosphate aldolase (Fba1) (An02g07470) of glycolysis, Nmt1 (An02g10320) for thiamine biosynthesis and Vip1 (An16g05020), an inositol phosphate kinase. The former two are the enzymes for carbon metabolism, and the latter is for the synthesis of inositol phosphates that play roles in signal transduction for processes such as cellular growth, apoptosis, and endocytosis. It is not possible to determine whether these proteins were absolutely reduced in the enzyme-producing strain or were simply diluted in the protein extract due to the presence of heterologous enzyme.
Figure 1. Visual comparison of 2D-gel electrophoresis with two strains. The spots shown in green are from C1100 producing a heterologous enzyme, and the spots shown in pink are from C1844 not producing enzyme. The samples collected at 48 h were used. The 1st dimensional electrophoresis was conducted at the gradient pH 4–7. The spots shown as HP are the ones from heterologous glucoamylase. ND is a protein not determined. The rest of proteins are described in the text. GpdA, Mdh1, HexA, Tpm1 were abundant, but did not appear in the gel of this pH range. These protein spots can be detected on the gels of pH 3–10 (Figure S1).

Quantitative comparison of detected proteins by measurement of spot-intensities
A quantitative analysis was undertaken to provide a more accurate comparison of protein levels secreted by the two strains. To conduct a quantitative comparison between the two strains, Coomassie brilliant blue-stained gels (large scale, pH 3–10) were imaged and spot intensities were measured by the image scanner. The intensity of each protein spot per the total intensity of all protein spots was calculated as a percentage (%). When a protein appeared as multiple spots on a gel, intensities of these spots were combined and used for the calculation. The results are shown in . As mentioned before, in the case of C1100, the produced heterologous glucoamylase appeared on the gels. The intensity of this enzyme was 16.5%, if included in the calculation of the total intensity of the gel. It was the most abundant protein in this sample, but, since it is not a native protein of A. niger, it was not included in the calculation of the total intensity of the spots on the gel. The 28 most abundant proteins in each strain are listed in .
Table 2. Comparison of the detected proteins on two strains with 48-h samples.
Analysis of increased abundance upon enzyme production
ER chaperones and the unfolded protein response (UPR)
Among the 28 most abundant proteins, 11 proteins were abundant especially in the enzyme-producing strain (). They were primarily chaperones and foldases, as mentioned in the previous section. Their elevated abundance will be discussed in the context of a potential role in enzyme production by this strain.
Table 3. Proteins abundant in the samples of 48 h.
BipA is an ER chaperone that assists in the translocation of newly synthesized proteins to the ER and their proper folding within the ER [Citation32]. BipA is also a sensor for unfolded proteins in the ER and interacts with Ire1 to activate the transcription factor HacA and induce the unfolded protein response (UPR) [Citation33]. PDI is a foldase that assists S-S bond formation for correct protein folding [Citation34] which is necessary for optimal secretion [Citation35]. Calnexin is an ER chaperone that binds only to N-linked glycoproteins [Citation36] and assists their proper folding. The heterologous glucoamylase was a heavily glycosylated protein. Therefore, calnexin was likely necessary for proper folding and secretion of this enzyme.
Over-expression of a secreted protein gives rise to secretion stress in the ER and leads to UPR [Citation10,Citation20,Citation37]. Both BipA and PDI [Citation38,Citation39], as well as calnexin [Citation20,Citation40], are known to be up-regulated by the UPR, which would fit with a hypothesis whereby over-expression of the heterologous enzyme in our strain induces this stress response. A previous transcriptomic analysis on an A. niger strain over-expressing a native glucoamylase [Citation40] identified elevated transcription of 130 genes linked to the secretory pathway. The genes were categorized into four functional groups; ER membrane translocation, protein glycosylation, vesicle transport, and ion homeostasis. Among these upregulated genes, besides BipA, PDI, and calnexin, only six also appeared in this study. Corresponding proteins detected in our study include two protein disulfide isomerases, TigA (An18g02020) and PrpA (An01g04600), the chaperone Lhs1 (An01g13220), the beta-subunit of ER alpha-glucosidase (An13g00620) linked to protein glycosylation, golgin-160 related protein Rud3 (An08g00290) (for Golgi transport), and a hypothetical protein YIL041w (An02g03460), with predicted function in the secretory pathway. No other detected proteins matched genes identified by transcriptomics. The reason for this discrepancy could be the difference in the tested strains, but it is also the case that results of transcriptome and proteome analyses are not well correlated [Citation10].
The UPR is closely related to ER-associated protein degradation (ERAD) [Citation25,Citation33,Citation41]. In this study, several proteins related to proteasome and ERAD were detected (Table S1). Of these, Cdc48 (An04g09170) is a chaperone with AAA+ ATPase activity involved in ERAD via the ubiquitin fusion degradation pathway [Citation42–Citation45]. In ERAD, Cdc48 recognizes misfolded proteins in the ER and translocates them to the cytoplasm for degradation by the proteasome [Citation44]. Calnexin is also known to play a role in ERAD [Citation46]. The identification of proteins involved in both the UPR and ERAD in our proteomic study indicates that both stress responses are occurring during heterologous enzyme production. However, the gene products of several major ERAD components, such as derA, doaA, hrdC, which are known to be up-regulated during the UPR [Citation25,Citation41], were not detected by our proteomics. Again, this might be attributable to the strain difference, or the difference between transcriptomic analysis and proteomic analysis. It would be interesting to conduct transcriptomic analysis on our strains to clarify it.
Potential repression by secretion stress upon enzyme over-production
The quantitative analysis enabled us to compare the protein population between the two strains. This comparison not only identified more abundant proteins, as described above, but also proteins that decreased in abundance in the enzyme-producing strain relative to the reference strain.
When the total intensity (%) of each spot was compared, enzymes in carbon metabolism, such as Fba1 and enolase, as well as Vip1, an inositol phosphate kinase, were detected at an over two-fold lower level in the enzyme-producing strain. It is, therefore, possible that these enzymes are downregulated in this enzyme-producing strain. It is known that the over-expression of a secreted protein causes ER stress and represses the expression of other secreted proteins [Citation47], a phenomenon called REpression under Secretion Stress (RESS) [Citation48]. Although RESS sounds a good explanation of the down-regulation of proteins observed here, it is a mechanism of transcriptional regulation and does not affect the expression of non-secreted protein [Citation47]. Carbon metabolizing enzymes are not secreted protein; thus, their observed repression in this study must be due to a different mechanism.
Other abundant proteins
In addition to the above-mentioned abundant proteins, linked to the UPR and ERAD pathways which might be expected due to known induction of such stress responses upon enzyme-production, other identified proteins of significant abundance in the test strain are worth discussion (see ). Such proteins include the house-keeping chaperones, Ssa4 (An07g09990) and Ssb2 (An16g09260), cAMP-dependent protein kinase (An16g03740), cytochrome C peroxidase (CCP) (04g04060), some more chaperones, and signaling proteins.
The cAMP-dependent protein kinase, or protein kinase A (PKA), plays important roles in the cAMP signaling cascade [Citation49] in response to environmental fluctuations. It evokes various responses related to cell development, metabolisms, and virulence, through its protein phosphorylation activity of a range of targets such as protein kinases, signaling factors, and transcription factors [Citation49,Citation50]. In Aspergillus, PKA is known to be involved in controlling morphology [Citation51], lipid composition [Citation52], conidial germination [Citation53], and virulence [Citation54]. Its direct or indirect phosphorylation targets are not well characterized. Recently, a comprehensive phosphorylome was conducted on A. nidulans [Citation50] and identified CreA, a main repressor for carbon catabolite repression, as one of the targets. PKA in A. niger may also be involved in sensing glucose and controlling glucose repression, besides controlling morphology.
Cytochrome C peroxidase (CCP) is a protein involved in the oxidative stress response by decreasing reactive oxygen species (ROS), such as hydrogen peroxide [Citation55]. It is known that ROS are continuously formed during aerobic metabolism [Citation56]. Moreover, it was suggested that active protein secretion caused oxidative stress [Citation10]. Previously reported proteome studies of A. niger also detected proteins that reduce ROS, such as superoxide dismutase, peroxiredoxin [Citation22], or thioredoxin [Citation23]. The detection of CCP in our study is thus consistent with these previous studies.
House-keeping chaperones, such as Ssa4, Ssb2, and Ssc1, are present abundantly in S. cerevisiae [Citation57]. These proteins were detected at a high level in our strains as well. The quantitative analysis revealed that these chaperones were at about a two-fold higher level in the enzyme-producing strain than the reference strain (data not shown).
Ssc1 is a mitochondrial chaperone that assists the translocation of mitochondrial proteins newly born in the cytosol [Citation58,Citation59]. Various cellular reactions relating to the energy generation, including the TCA cycle, pyruvate metabolism, as well as the electron transport system, occur in the mitochondria; hence, Ssc1 is likely required at a high level to support these cellular reactions.
Ssb2 is primarily a ribosome-binding chaperone located in the cytosol [Citation57,Citation60] that acts against nascent polypeptides [Citation61] to prevent aggregation and potential interaction with the signal recognition particle (SRP) which would miss-target them to the ER. Ssb2 also has a role in ribosome-independent glucose sensing [Citation62], controlling the activity of protein kinase Snf1, a central player in glucose repression via interaction with signal transduction protein(s). In yeast, 14-3-3 proteins, Bmh1 and Bmh2, are involved in glucose sensing jointly with Ssb [Citation63,Citation64]. In A. nidulans, a homolog ArtA is involved in septum formation and conidia polarization [Citation65]. 14-3-3 homologs, ArtA (An18g06270) and ArtB (An07g07760), were detected at a high level in our strains, which could have roles in glucose sensing and repression in A. niger.
Ssa4 is a cytosol chaperone, and in yeast, it serves to protect cells from stressors, such as heat, starvation, and oxidants [Citation66,Citation67]. Ssa4 shuttles between the nucleus and the cytosol during normal growth conditions, but remains in the nucleus under stressed conditions [Citation68]. Sbp1 (An18g02140), which was also detected in abundance, is linked to the activity of Ssa4 in nuclear export. Data suggest that Ssa4 actively increases in A. niger during fermentation.
Proteins abundant in both strains
Although proteins abundant in both strains may not be as interesting from the point of view of heterologous enzyme production in A. niger and a better understanding of how cellular activity is affected by enzyme production, it is worth highlighting some of those detected since so many A. niger genes have not been characterized.
Metabolic enzymes
Among the 28 most abundant proteins, 17 proteins appeared in both strains (), indicating they have important roles in overall cell growth and function rather than enzyme production. The most dominant functional group of the category comprises enzymes related to carbon metabolism. As mentioned previously, glycolytic enzymes and Nmt1 were detected at over two-fold lower levels in the enzyme-producing strains, although they were still abundant. The quantities of mAspAT and transaldolase (Tal1) remained equally or slightly higher in the enzyme-producing strains. Together, the analysis suggests that the metabolic pathway to gain energy shifted from glycolysis to other pathways, such as the pentose phosphate pathway, during enzyme over-production.
Signaling proteins
There were a couple of proteins abundant in both strains which are likely involved in signal transduction or regulation of cellular activities. These include a hypothetical translationally controlled tumor protein (TCTP) (An01g10050), two 14-3-3 family proteins ArtA and ArtB, a hypothetical protein with a predicted Pleckstrin-homology (PH) domain (An01g07870), and a newly identified hypothetical protein (An09g06410), CSAP. Although abundant in both strains, the quantitative analysis indicated that they were slightly elevated in the enzyme-producing strains.
TCTP is a small multi-functional protein of around 20 kDa, and highly conserved and ubiquitous in eukaryote cells [Citation69]. It is likely involved in cell growth, cell cycling, and protecting cell against various stressors and apoptosis [Citation70,Citation71]. A homolog in A. nidulans, TcpA, was involved in the branching of vegetative mycelia and in the development of asexual spores [Citation72]; however, it is unlikely that these functions are important during the submerged fermentation for enzyme production. The role of a TCTP homolog found in A. niger, termed AngA, could be protecting cells from stressors during fermentation or supporting cell growth other than branching.
A newly identified hypothetical protein (An09g06410) was abundant in both strains. The protein is relatively small, consisting of 256 amino acid residues, with multiple repeats of glutamine, glycine, and serine. C-terminal half of the protein has a similarity to WW domain proteins such as A. fumigatus Afu5g03750 (Figure S2) or A. flavus RMZ45354. WW domain is a highly conserved protein domain involved in protein–protein interaction [Citation73]. It appears in various proteins working for signal transduction. There are a few orthologues in A. nidulans (AN9521, AN0790), A. fumigatus (Afu5g03750), A. oryzae (AO090102000585), and other Aspergillus species. Therefore, this protein is likely a conserved protein among Aspergillus. A protein-protein BLAST search using the NCBI database did not yield any hits from S. cerevisiae, in support of the proposition that it is a unique protein for fungal species. Here we term the protein as Conserved Small Abundant Protein in Aspergillus (CSAP). The function of CSAP is yet to be elucidated.
Another hypothetical protein (An01g07870), termed Aph1, had a predicted Pleckstrin-homology (PH) domain [Citation74], which is known to bind phosphorylated phosphatidylinositol, a lipid mediator located on the membrane of the organelle. The domain recruits the protein to the targeted organelle for signal transduction. As orthologues exist in A. oryzae (AO090009000457), A. fumigatus (Afu4g12450), A. nidulans (AN3674) and other Aspergillus species (data not shown), it is likely a conserved protein among Aspergillus. Exact roles for this protein in A. niger remain to be determined.
Cell components – HexA and tropomyosin
Among the detected cell components, the most abundant protein was HexA (An07g04570), which is a core protein of the Woronin body [Citation75]. Similar results were observed in a Trichoderma reesei strain producing cellulases [Citation76], suggesting the importance of this protein during enzyme-production. The Woronin body functions to seal septal pores upon cellular damage to avoid cell lysis [Citation77]. Observations show that fungal cells might be damaged during fermentation; hence, HexA was required in high amounts to prevent mycelial death. Alternatively, it was observed in N. crassa that the expression of HexA was maximal in apical cells [Citation78]. It is also possible that the high level of HexA in this study may be related to active apical cell growth during fermentation.
Another abundant cellular protein was tropomyosin (An13g00760), which is a conserved actin-binding protein that stabilizes actin filaments [Citation79]. Tropomyosin particularly has a high affinity to actin cable, so that it is used as a marker for it [Citation80,Citation81]. Actin cable plays a role in apical exocytosis assisting tip growth. In fact, tropomyosin was observed in the Spitzenkörper at the hyphal tip, but is also concentrated near the developing septa [Citation82]. Importantly, protein secretion typically occurs in the apical cells [Citation83] but can also occur near the septum [Citation84]. An abundance of tropomyosin may be related to the stabilization of actin cable to support active secretion from the hyphal tip and septum. To our knowledge, our study represents the first discussion of tropomyosin function in A. niger.
Retention of secreted over-produced enzyme within the cells
One of the striking observations in this proteomic study was that the heterologous glucoamylase, expressed for secretion outside of the cells, appeared in the proteome (). Whether this glucoamylase accumulated inside the cell or associated with the cell wall, similarly to an alpha-amylase in A. oryzae [Citation85], was not known. However, the concentration of the observed enzyme in the proteome was higher later stage of fermentation, indicating that accumulation is occurring during fermentation. Interestingly, the size of abundant spots for glucoamylase varied during fermentation, smaller sizes appearing with higher frequency in the 120-h sample (). This suggests that the enzyme protein probably does accumulate inside the cell, and is subject to proteolytic digestion during fermentation. Besides the detection of proteins related to the proteasome, a couple of vacuolar proteases, such as pepE, pepC and carboxypeptidase Y, was detected in the proteome. Whether glucoamylase was degraded by the proteasome via ERAD, or digested by vacuolar degradation cannot be established in this study.
Figure 2. Accumulation of glucoamylase in the cells. (a) Three 2D-gels of C1100 at different time points. Spots shown by arrows were originated from glucoamylase. Population of spots increased with time, indicating produced glucoamylase was being accumulated in the cells. (b) Magnified 2D-gel of the C1100 collected at 120 h. Spots shown with arrows had partial sequence of glucoamylase. This indicates the produced glucoamylase was digested in the cells.

Observation of intracellular glucoamylase was surprising, as the production of the enzyme in the culture broth increased linearly (data not shown). It is suggestive of most of the enzyme having been secreted, with a fraction remaining in the cell. The cause of retention could be indicative of a malfunction in a part of secretion machinery for newly borne protein, such as ER-entering, protein folding, glycosylation, and/or vesicle-trafficking, or could indicate over-capacity of the secretion pathway. Previously, glucoamylase was detected in the A.niger proteome, but predominantly as a protein in the secretome [Citation24]. In that case, the detected protein was likely an intermediate of a successful secretion process. Our observation is probably the first evidence of the accumulation of a proportion of over-secreted enzyme in the cells during fermentation. Whatever the cause of this observation, our results indicate that there is some room for improvement of heterologous protein production.
Conclusion
Simple proteomic analysis of industrial A. niger strains producing enzyme suggested that the UPR was happening, with the evidence that BipA, PDI, and calnexin, all known as UPR elements, were detected at a high level in the enzyme-producing strain. It was probably due to the secretion stress caused by over-expression of a heterologous enzyme, as already reported by several researchers. A chaperone Cdc48, linked to ERAD, was also more abundant, that suggests that ERAD was also occurring. However, other known components identified by the transcriptomic analysis, which were up-regulated in the UPR and ERAD in A. niger, were not observed in our proteomic study (with exception of six proteins in addition to the above). Additional investigations would be needed to clarify the reason for this discrepancy.
Besides, there were a couple of new findings through this study.
It was observed that enzymes in carbon metabolisms and others, such as inositol-phosphate kinase, were significantly reduced in the enzyme-producing strain. Their spot-intensities were at over two-fold lower level compared to the reference. The results indicated the shift of metabolic pathway to other than glycolysis. The phenomenon sounds similar to a known protein expression repression, repression under secretion stress (RESS); however, RESS affects the expression of secreted proteins only; therefore, the observed phenomenon is different from RESS and must be due to another mechanism.
A couple of conserved proteins, likely playing roles in signal transduction or regulation of cellular activities, was relatively abundant regardless of the enzyme production. This time, two new components, termed CSAP (Conserved Small Abundant Protein in Aspergillus) and Aph1, were identified as abundant components in our strains and conserved among fungal strains. The roles of these proteins have not been elucidated and awaits further study.
HexA, a core protein of Woronin body, and tropomyosin, which has a high affinity to actin cable, were detected at a high level from both, a tested and the reference strains. They must play important roles during fermentation, such as protecting cells or supporting apical growth, or assisting exocytosis for active secretion.
There was a surprising observation, but it was found that a portion of the expressed secreted-enzyme remained inside the cells and accumulated during enzyme production. Moreover, there is evidence that this accumulated enzyme protein was digested during the fermentation. The results indicate the mal-function of the secretion pathway in the enzyme-producing strain, which might be due to the over-expression of heterologous enzyme. It suggests that improvement of the secretion machinery in our strain could improve the enzyme-production yield even further. Whether the mechanism of digestion was ERAD or another mechanism was outside the scope of this study.
Contribution of authors
ST initiated the study and conducted the analysis and interpretation of the results. KK prepared biological samples and the initial dataset with annotation for the analysis. SO conducted the proteomes including the search for protein identification and measurement of spot intensity.
Supplement-v2-f.pdf
Download PDF (719.7 KB)Acknowledgments
We thank Mayuko Kodama for the technical support, Michiko Ihara and Hiroaki Udagawa for the analytical support, and Carsten Hjort for the approval of the publication of this study. We would like to thank Enago (www.enago.jp) for the English language review.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
References
- Schuster E, Dunn-Coleman N, Frisvad J, et al. On the safety of Aspergillus niger - a review. Appl Microbiol Biotechnol. 2002;59(4–5):426–435.
- van Dijck PWM, Selten GCM, Hempenius RA. On the safety of a new generation of DSM Aspergillus niger enzyme production strains. Regul Toxicol Pharmacol. 2003;38(1):27–35.
- Andersen MR, Salazar MP, Schaap PJ, et al. Comparative genomics of citric-acid-producing Aspergillus niger ATCC 1015 versus enzyme-producing CBS 513.88. Genome Res. 2011;21(6):885–897.
- de Vries RP, Visser J. Aspergillus enzymes involved in degradation of plant cell wall polysaccharides. Microbiol Mol Biol Rev. 2001 Dec;65(4):497–522.
- Berka RM, Dunn-Coleman N, Ward M. Industrial enzymes from Aspergillus species. In: Bennett JW, Klich MA, editors. Aspergillus, biology and industrial applications. Stoneham (MA): Butterworth-Heinemann; 1992. p. 155–202.
- Lubertozzi D, Keasling JD. Developing Aspergillus as a host for heterologous expression. Biotechnol Adv. 2009;27(8):53–75.
- Gouka RJ, Punt PJ, van den Hondel CA Efficient production of secreted proteins by Aspergillus: progress, limitations and prospects. Appl Microbiol Biotechnol. 1997;47(1):1–11.
- Punt PJ, van Biezen N, Conesa A, et al. Filamentous fungi as cell factories for heterologous protein production. Trends Biotechnol. 2002 May;20(5):200–206.
- Fleer A, Dersch P. Expression and export: recombinant protein production systems for Aspergillus. Appl Microbiol Biotechnol. 2010;87(4):1255–1270.
- Jacobs DI, Olsthoorn MM, Maillet I, et al. Effective lead selection for improved protein production in Aspergillus niger based on integrated genomics. Fungal Genet Biol. 2009;46(1):141–152.
- Lombrana M, Moralejo FJ, Pinto R, et al. Modulation of Aspergillus awamori thaumatin secretion by modification of bipA gene expression. Appl Environ Microbiol. 2004;70(9):5145–5152.
- Carvalho ND, Arentshorst M, Kooistra R, et al. Effects of a defective ERAD pathway on growth and heterologous protein production in Aspergillus niger. Appl Microbiol Biotechnol. 2011;89(1):357–373.
- Fiedler MRM, Barthel L, Kubisch C, et al. Construction of an improved Aspergillus niger platform for enhanced glucoamylase secretion. Microb Cell Fact. 2018;17(1):95.
- Meyer V, Wu B, Ram AFJ. Aspergillus as a multi-purpose cell factory: current status and perspectives. Biotechnol Lett. 2011;33(3):469–476.
- Liu L, Feizi A, Österlund T, et al. Genome-scale analysis of the high-efficient protein secretion system of Aspergillus oryzae. BMC Syst Biol. 2014;8(16):73.
- Knuf C, Nielsen J. Aspergilli: systems biology and industrial applications. Biotechnol J. 2012;7(9):1147–1155.
- Andersen MR, Nielsen J. Current status of systems biology in Aspergilli. Fungal Genet Biol. 2009;46:180–190.
- Andersen M, Nielsen M, Nielsen J. Metabolic model integration of the bibliome, genome, metabolome and reactome of Aspergillus niger. Mol Syst Biol. 2008;4(1):78.
- Pel H, de Winde J, Archer D, et al. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat Biotechnol. 2007;25(2):221–231.
- Guillemette T, van Peij NN, Goosen T, et al. Genomic analysis of the secretion stress response in the enzyme-producing cell factory Aspergillus niger. BMC Genomics. 2007;8(1):158.
- Jorgensen TR, Goosen T, van den Hondel CA, et al. Transcriptomic comparison of Aspergillus niger growing on two different sugars reveals coordinated regulation of the secretory pathway. BMC Genomics. 2009;10(1):44.
- Lu X, Sun J, Nimtz M, et al. The intra- and extracellular proteome of Aspergillus niger growing on defined medium with xylose or maltose as carbon substrate. Microb Cell Fact. 2010;9(17):23.
- Miguel J, de Oliveira PF, van Passel MWJ, et al. Shotgun proteomics of Aspergillus niger microsomes upon D-Xylose induction. Appl Environ Microbiol. 2010 July;76(13):4421–4429.
- Miguel J, de Oliveira PF, van Passel MWJ, et al. Proteomic analysis of the secretory response of Aspergillus niger to D-Maltose and D-Xylose. PLoS ONE. 2011;6(6):e20865.
- Carvalho NDSP, Jorgensen TR, Arentshorst M, et al. Genome-wide expression analysis upon constitutive activation of the HacA bZIP transcription factor in Aspergillus niger reveals a coordinated cellular response to counteract ER stress. BMC Genomics. 2012;13(1):350–366.
- Lu H, Cao W, Liu X, et al. Multi-omics integrative analysis with genome-scale metabolic model simulation reveals global cellular adaptation of Aspergillus niger under industrial enzyme production condition. Sci Rep. 2018;8(16):14404.
- Sloothaak J, Odoni DI, de Graaff LH, et al. Aspergillus niger membrane-associated proteome analysis for the identification of glucose transporters. Biotechnol Biofuels. 2015;8(1):150.
- Udagawa H, Landvik S, Ihara M, et al. Inventors; Novozymes A/S, Novozymes North America, Inc. assignee. Polypeptides having glucoamylase activity and polynucleotides encoding the same. US patent 8148127 B2. 2012 Apr 3.
- Udagawa H Inventor; Novozymes A/S, assignee. Simultaneous site-specific integrations of multiple gene-copies in filamentous fungi. 2012 Nov. 29. PCT application WO2012/160093.
- Pedersen H, Beyer M, Nielsen J. Glucoamylase production in batch, chemostat and fed-batch cultivations by an industrial strain of Aspergillus niger. Appl Microbiol Biotechnol. 2000;53(3):272–277.
- Tatusov RL, Fedorova ND, Jackson JD, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003 Sep 11;4(1):1–4. (KOG).
- van Gemeren IA, Punt PJ, Drint-Kuyvenhoven A, et al. The ER chaperone encoding bipA gene of black Aspergilli is induced by heat shock and unfolded proteins. Gene. 1997 Oct;198(1–2):43–52.
- Mori K. Signaling pathways in the unfolded protein response: development from yeast to mammals. J Biochem. 2009;146(6):743–750.
- Ngiam C, Jeenes DJ, Punt PJ, et al. Characterization of a foldase, protein disulfide isomerase A, in the protein secretory pathway of Aspergillus niger. Appl Environ Microbiol. 2000;66(2):775–782.
- Moralejo FJ, Watson AJ, Jeenes DJ, et al. A defined level of protein disulfide isomerase expression is required for optimal secretion of thaumatin by Aspergillus awamori. Mol Genet Enom. 2001;266:245–253.
- Zapun A, Darby NJ, Tessier DC, et al. Enhanced catalysis of ribonuclease B folding by the interaction of calnexin or calreticulin with ERp57. J Biol Chem. 1998;273(11):6009–6012.
- Schroder M, Kaufman RJ. ER stress and the unfolded protein response. Mutat Res. 2005;569(1–2):29–63.
- Sims AH, Gent ME, Lanthaler K, et al. Transcriptome analysis of recombinant protein secretion by Aspergillus nidulans and the unfolded-protein response in vivo. Appl Environ Microbiol. 2005 May;71(5):2737–2747.
- Mulder HJ, Saloheimo M, Penttilä M, et al. The transcription factor HACA mediates the unfolded protein response in Aspergillus niger, and up-regulates its own transcription. Mol Gen Genet. 2004;271(2):130–140.
- Kwon MJ, Jogensen TR, Nitsche BM, et al. The transcriptomic fingerprint of glucoamylase over-expression in Aspergillus niger. BMC Genomics. 2012;13(1):701.
- Travers KJ, Patil CK, Wodicka L, et al. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000 Apr 28;101(3):249–258.
- Bodnar N1, Rapoport T1. Toward an understanding of the Cdc48/p97 ATPase. F1000Res. 2017 Aug 3;6:1318.
- Ye Y, Meyer HH, Rapoport TA. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414(6864):652–656.
- Jarosch E, Taxis C, Volkwein C, et al. Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nat Cell Biol. 2002;4(2):134–139.
- Wu X, Rapoport TA. Mechanistic insights into ER-associated protein degradation. Curr Opin Cell Biol. 2018 Aug;53:22–28.
- Hoseki J, Ushioda R, Nagata K. Mechanism and components of endoplasmic reticulum-associated degradation. J Biochem. 2010;147(1):19–25.
- Al-Sheikh H, Watson AJ, Lacey GA, et al. Endoplasmic reticulum stress leads to the selective transcriptional downregulation of the glucoamylase gene in Aspergillus niger. Mol Microbiol. 2004;53(6):1731–1742.
- Pakula TM, Laxell M, Huuskonen A, et al. The Effects of drugs inhibiting protein secretion in the filamentous fungus Trichoderma reesei. J Biol Chem. 2003;278(45):45011–45020.
- Oliver BG, Panepinto JC, Fortwendel JR, et al. Cloning and expression of pkaC and pkaR, the genes encoding the cAMP-dependent protein kinase of Aspergillus fumigatus. Mycopathologia. 2002;154(2):85–91.
- Ribeiro LFC, Chelius C, Boppidi KR, et al. Comprehensive analysis of Aspergillus nidulans PKA phosphorylome identifies a novel mode of CreA regulation. mBio. 2019 Apr 30;10(2). DOI:10.1128/mBio.02825-18
- Saudohar M, Bencina M, van de Vondervoort PJI, et al. Cyclic AMP-dependent protein kinase is involved in morphogenesis of Aspergillus niger. Microbiology. 2002 Aug;148(Pt 8):2635–2645.
- Jernejc K, Bencina M. Lipid composition of cAMP-dependent protein kinase mutants of Aspergillus niger. FEMS Microbiol Lett. 2003 Aug;225(2):291–297.
- Fuller KK, Richie DL, Feng X, et al. Divergent protein Kinase A isoforms co-ordinately regulate conidial germination, carbohydrate metabolism and virulence in Aspergillus fumigatus. Mol Microbiol. 2011;79(4):1045–1062.
- Liebmann B, Müller M, Braun A, et al. The cyclic AMP-Dependent protein Kinase A network regulates development and virulence in Aspergillus fumigatus. Infect Immun. 2004;72(9):5193–5203.
- Shin Y, Lee S, Ku M, et al. Cytochrome c peroxidase regulates intracellular reactive oxygen species and methylglyoxal via enzyme activities of erythroascorbate peroxidase and glutathione-related enzymes in Candida albicans. Int J Biochem Cell Biol. 2017 Nov;92:183–201.
- Keiner M, Harvey LM, McNeil B. Morphological and enzymatic responses of a recombinant Aspergillus niger to oxidative stressors in chemostat cultures. J Biotechnol. 2003;100(3):251–260.
- Peisker K, Chiabudini M, Rospert S. The ribosome-bound Hsp70 homolog Ssb of Saccharomyces cerevisiae. Biochim Biophys Acta. 2010;1803(6):662–672.
- Kang P-J, Ostermann J, Shilling J, et al. Requirement for hsp70 in the mitochondrial matrix for translocation and folding of precursor proteins. Nature. 1990;348(6297):137–143.
- Liu Q, Krzewska J, Liberek K, et al. Mitochondrial Hsp70 Ssc1: role in protein folding. J Biol Chem. 2000;276(9):6112–6118.
- Gumiero A, Conz C, Valentín G, et al. Interaction of the cotranslational Hsp70 Ssb with ribosomal proteins and rRNA depends on its lid domain. Nat Commun. 2016;7(1):13563.
- Döring K, Ahmed N, Riemer T, et al. Profiling Ssb-Nascent chain interactions reveals principles of Hsp70-assisted folding. Cell. 2017 Jul 13;170(2):298–311.
- von Plehwe U, Berndt U, Conz C, et al. The Hsp70 homolog Ssb is essential for glucose sensing via the SNF1 kinase network. Genes Dev. 2009 Sep 1;23(17):2102–2115.
- Hübscher V, Mudholkar K, Chiabudini M, et al. The Hsp70 homolog Ssb and the 14-3-3 protein Bmh1 jointly regulate transcription of glucose repressed genes in Saccharomyces cerevisiae. Nucleic Acids Res. 2016 Jul;44(12):5629–5645.
- Dombek KM, Kacherovsky N, Young ET. The Reg1-interacting proteins, Bmh1, Bmh2, Ssb1, and Ssb2, have roles in maintaining glucose repression in Saccharomyces cerevisiae. J Biol Chem. 2004 Sep 10;279(37):39165–39174.
- Kraus PR, Hofmann AF, Harris SD. Characterization of the Aspergillus nidulans 14-3-3 homologue, ArtA. FEMS Microbiol Lett. 2002 Apr 23;210(1):61–66.
- Chughtai ZS, Rassadi R, Matusiewicz N, et al. Starvation promotes nuclear accumulation of the hsp70 Ssa4p in yeast cells. J Biol Chem. 2001 Jun;276(23):20261–20266.
- Quan X, Rassadi R, Rabie B, et al. Regulated nuclear accumulation of the yeast hsp70 Ssa4p in ethanol-stressed cells is mediated by the N-terminal domain, requires the nuclear carrier Nmd5p and protein kinase C. Faseb J. 2004 May;18(7):899–901.
- Quan XX, Tsoulos P, Kuritzky A, et al. The carrier Msn5p/KAp142p promotes nuclear export of the hsp70 Ssa4p and relocates in response to stress. Mol Microbiol. 2006;62(2):592–609.
- Bommer UA, Thiele BJ. The translationally controlled tumour protein (TCTP). Int J Biochem Cell Biol. 2001 Mar;36(3):379–385.
- Rinnerthaler M, Lejskova R, Grousl T, et al. Mmi1, the yeast homologue of mammalian TCTP, associates with stress granules in heat-shocked cells and modulates proteasome activity. PLoS One. 2013 Oct;8(10):e77791.
- Vojtova J, Hasek J. Mmi1, the yeast ortholog of mammalian translationally controlled tumor protein (TCTP), negatively affects rapamycin-induced autophagy in post-diauxic growth phase. Cells. 2020 Jan;9(1):E138.
- Oh YT, Ahn CS, Jeong YJ, et al. Aspergillus nidulans translationally controlled tumor protein has a role in the balance between asexual and sexual differentiation and normal hyphal branching. FEMS Microbiol Lett. 2013 Jun;343(1):20–25.
- Hesselberth JR, Miller JP, Golob A, et al. Comparative analysis of saccharomyces cerevisiae WW domains and their interacting proteins. Genome Biol. 2006;7(4):R30.
- Musacchio A, Gibson T, Rice P, et al. The PH domain: a common piece in the structural patchwork of signaling proteins. Trends Biochem Sci. 1993 Sep;18(9):343–348.
- Dhavale T, Jedd G. The fungal Woronin body. In: Howard RJ, Gow NAR, editors. Biology of the fungal cell. 2nd ed. Berlin: Springer; 2007. p. 87–94.
- Lim D, Hains P, Walsh B, et al. Proteins associated with the cell envelope of Trichoderma reesei: a proteomic approach. Proteomics. 2001;1:899–910.
- Maruyama J, Juvvadi PR, Ishi K, et al. Three-dimensional image analysis of plugging at the septal pore by Woronin body during hypotonic shock inducing hyphal tip bursting in the filamentous fungus Aspergillus oryzae. Biochem Biophys Res Commun. 2005;331(4):1081–1088.
- Tey WK, North AJ, Peyes JL, et al. Polarized gene expression determines Woronin body formation at the leading edge of the fungal colony. Mol Biol Cell. 2005 June;16(6):2651–2659.
- Gunning PW, Hardeman EC, Lappalainen P, et al. Tropomyosin - master regulator of actin filament function in the cytoskeleton. J Cell Sci. 2015 Aug;128(16):2965–2974.
- Bergs A, Ishitsuka Y, Evangelinosa M, et al. Dynamics of actin cables in polarized growth of the filamentous fungus Aspergillus nidulans. Front Microbiol. 2016;7:682.
- Takeshita N, Evangelinosa M, Zhoud L, et al. Pulses of Ca2+ coordinate actin assembly and exocytosis for stepwise cell extension. PNAS. 2017 May;114(22):5701–5706.
- Taheri-Talesh N, Horio T, Araujo-Baza´n L, et al. The tip growth apparatus of Aspergillus nidulans. Mol Biol Cell. 2008 April;19(4):1439–1449.
- Gordon CL, Archer DB, Jeenes DJ, et al. A glucoamylase: GFP gene fusion to study protein secretion by individual hyphae of Aspergillus niger. J Microbiol Methods. 2006;42(1):39–48.
- Hayakawa Y, Ishikawa E, Shoji J, et al. Septum-directed secretion in the filamentous fungus Aspergillus oryzae. Mol Microbiol. 2011;81(1):40–55.
- Zhang S, Sato H, Ichinose S, et al. Cell wall α-1,3-glucan prevents α-amylase adsorption onto fungal cell in submerged culture of Aspergillus oryzae. J Biosci Bioeng. 2017;124(1):47–53.
