ABSTRACT
Polymorphism of the prion protein gene (PRNP) gene determines an animal’s susceptibility to scrapie. Three polymorphisms at codons 136, 154, and 171 have been linked to classical scrapie susceptibility, although many variants of PRNP have been reported. However, no study has investigated scrapie susceptibility in Nigerian sheep from the drier agro-climate zones. In this study, we aimed to identify PRNP polymorphism in nucleotide sequences of 126 Nigerian sheep by comparing them with public available studies on scrapie-affected sheep. Further, we deployed Polyphen-2, PROVEAN, and AMYCO analyses to determine the structure changes produced by the non-synonymous SNPs. Nineteen (19) SNPs were found in Nigerian sheep with 14 being non-synonymous. Interestingly, one novel SNP (T718C) was identified. There was a significant difference (P < 0.05) in the allele frequencies of PRNP codon 154 between sheep in Italy and Nigeria. Based on the prediction by Polyphen-2, R154H was probably damaging while H171Q was benign. Contrarily, all SNPs were neutral via PROVEAN analysis while two haplotypes (HYKK and HDKK) had similar amyloid propensity of PRNP with resistance haplotype in Nigerian sheep. Our study provides valuable information that could be possibly adopted in programs targeted at breeding for scrapie resistance in sheep from tropical regions.
Introduction
Prion proteins (PRNP) play a crucial role in Transmissible Spongiform Encephalopathies (TSEs), which are distinctive diseases that can be inherited and contagious [Citation1–4]. In domestic animals, TSEs include scrapie in ovine and caprine, bovine spongiform encephalopathy (BSE) in cattle, and feline spongiform encephalopathy (FSE) in cats etc [Citation5–12]. The epidemic form of prion protein (PrPSc) has diverse structural dynamics compared to cellular prion (PrPC), and specific points of prion protein were found related to the conversion of PrCP to PrPSc [Citation13–16].
The susceptibility of animals to scrapie is predominantly controlled by polymorphism in the PRNP gene [Citation17]. Based on this, different single nucleotide mutations have been identified at codons 136 (A > V), 154 (R > H), and 171 (R > Q/H) of PRNP gene [Citation18,Citation19]. Amino acids at codons 141 and 154 have been found to be associated with various forms of classical scrapie through modification of the configuration of prion protein [Citation20,Citation21]. Changes at codon 136 from A to V has been reported to increase susceptibility to scrapie while variations at codon 171 from Q to R cause resistance in sheep [Citation18,Citation22].
It was believed that the ancestral PRNP gene was ARQ/ARQ (ARQ/ARQ wild type) in sheep [Citation23]. There are six primary forms of the wild-type allele which include ARQ, VRQ, AHQ, ARH, ARK, and ARR [Citation24]. Also, the genotypes associated with the admixture of these alleles are classified into five groups based on their degrees of resistance to scrapie [Citation25]. Recent studies have shown that amino acid substitution in some nucleotide positions were associated with resistance to scrapie and TSE in ARQ/ARQ sheep [Citation26].
Several studies have reported the polymorphism of the PRNP gene in sheep of different breeds or economic traits across different countries such as in testis-specific PRNP gene related to phenotypes of Chinese and Mongolian sheep [Citation27], phenotypic traits in different breeds of Chinese sheep [Citation28], Xinjiang local breeds in China [Citation29], and Tan sheep of Ningxia, China [Citation30]. In addition, studies on PRNP gene have been carried considering different traits such as milk traits in Latxa dairy [Citation31], reproductive traits in breeds of German sheep [Citation32], Baltic breeds [Citation33], pathogenesis and neuropathological phenotype [Citation34], Rasa Aragonesa (Spanish breed) [Citation7], milk production traits in Spanish Churra [Citation35], Greece [Citation36], Ireland [Citation37], Brazil [Citation38], Portugal [Citation39], Italy [Citation40], Turkey [Citation41,Citation42], and Palestine native sheep breed [Citation42]. Moreover, polymorphism studies of the PRNP gene in sheep from Inner Mongolian [Citation43] and Northwest China [Citation44] have been reported. In Africa, only few polymorphism studies on PRNP gene in sheep have been reported in Algeria [Citation45], Ethiopia [Citation46], Morocco [Citation47], and four Western African sheep population (i.e. Burkina-Sahel, Djallonke, Mossi and Touareg) [Citation48].
Nigerian sheep are reared in the drier agro-climatic zones of the country with an estimated population of 27 million [Citation49]. There are four major breeds of Nigerian sheep: Yankasa, Uda, Balami, and West Africa Dwarf [Citation50]. Sheep have socio-economic values in Nigeria as they form part of the livelihoods of small ruminant farmers that are majorly rural dwellers [Citation51]. Environmental factors of Nigeria impose health-related risks on the lives of human and animals which require urgent attention [Citation52].
Several studies have been carried out on Nigerian sheep using genetic techniques such as microsatellite DNA polymorphism [Citation53–56], polymorphism study on Myostatin (MSTN) [Citation57], and transferrin and haemoglobin [Citation58]. The survey conducted on the occurrence of scrapie in Jos within the central part of Nigeria [Citation59] is not sufficient to conclude on the absence of scrapie in Nigerian sheep. There is no report on polymorphism of the PRNP gene in Nigerian sheep, despite its significant effects on scrapie, the economic importance of Nigerian sheep, and transmission of the diseases from animals to humans.
Herein, we examined the genotype and allele frequencies of PRNP polymorphisms in 126 Nigerian sheep and compared the sampled population with previous studies on scrapie-affected animals in different breeds of sheep. Subsequently, we evaluated the linkage disequilibrium (LD) and analysed haplotypes of the PRNP polymorphisms in Nigerian sheep. Finally, we computed the biological impact, which includes the protein structure and functions of nonsynonymous SNPs via PolyPhen-2, PROVEAN, and AMYCO investigations.
Materials and methods
Blood sample collection and DNA extraction
We collected 10 ml of blood samples from 126 sheep (61 females and 65 males) from four different states in Nigeria namely, Kaduna State (n = 18 female; n = 15 male), Katsina State (n = 7 female; n = 8 male), Sokoto State (n = 9 female; n = 15 male), and Taraba State (n = 26 female; n = 28 male) (Supplementary Fig S1). During sample collection, we excluded sheep with close relationships after obtaining information from the herders. The whole blood samples were stored at −20 ◦C prior to DNA extraction. Genomic DNA was extracted at Kunming Institute of Zoology, Chinese Academy of Sciences (CAS) using the phenol-chloroform method [Citation60]. This was followed by the quantification using the Thermo Scientific™ NanoDrop 2000 spectrophotometer to evaluate the purity of the obtained DNA. Further, the quality of the total genomic DNA was checked by running gel electrophoresis using a 2% agarose gel with a 2 Kilobase (kb) DNA ladder marker. The 126 Nigerian sheep samples were then sequenced using the Sanger method.
Polymerase chain reaction (PCR) and DNA sequencing
To detect the polymorphic sites of the PRNP gene in sheep. PCR was performed with one pair of primers: PRNP-Forward (5’- CATTTATGACCTAGAATGTTTATAGCTGATGCCA −3’) and PRNP-Reverse (5’- TTGAATGAATATTATGTGGCCTCCTTCCAGAC −3’) [Citation45]. The 25 µl PCR mixture and sequencing reactions contained 1 μl of genomic DNA, 10 pmol of each primer, 2.5 mM dNTPs, and 5 units of Takara Taq DNA polymerase in a 10 pmol reaction buffer containing 1.5 mM MgCl2.
The PCR condition was as follows: 96°C for 5 min, 35 cycles of denaturation at 96°C for 30 s, 57°C for 15 s, 72°C for 1 min 30 s, and final extension of 72°C at 4 min. PCR products were purified for sequencing analysis with a QIAquick Gel Extraction Kit (Qiagen, Valencia California, USA). The PCR products were bidirectionally sequenced in an ABI 3730×L sequencer (Applied Biosystems, Foster City, California, USA).
Statistical analysis
Hardy Weinberg Equilibrium (HWE), Linkage Disequilibrium (LD) and haplotype distributions of the PRNP gene in Nigeria sheep were performed using DNA SNP Version 6.12.03 [Citation61]. The genotype differences, allele, and haplotype frequencies of the PRNP gene were analysed by chi-square test (χ2) or Fisher’s exact test using SPSS v21.0.
Evaluation of nonsynonymous SNPs in the caprine prion protein
Fourteen (14) nonsynonymous SNPs of the PRNP gene were evaluated using PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/) and PROVEAN (http://provean.jcvi.org/index.php). Furthermore, we determined the Amyloid propensity of ovine prion protein using alleles of PRNP SNPs via AMYCO (http://bioinf.uab.cat/amyco) [Citation62]. AMYCO is the algorithm for predicting amyloid fibril propensity from amino acid sequences.
Results
Identification of polymorphic sites of the PRNP gene in 126 Nigerian sheep
To achieve this, the open reading frame (ORF) of the PRNP gene in 126 Nigerian sheep was sequenced. The ORF contained 771 bp in length and occupied similar position with the PRNP gene of Ovis aries retrieved from the NCBI database (Gene ID:EF153678). In this study, we identified 19 SNPs, including 14 nonsynonymous SNPs being the T718C a novel SNP. (Supplementary Table S1 and Supplementary Figure S2). shows the genotype and allele frequencies of the 13 non-synonymous SNPs of PRNP in Nigerian sheep. Interestingly, all genotype frequencies of the identified SNPs conform to Hardy-Weinberg Equilibrium (HWE).
Table 1. Genotype and allele frequencies of 19 PRNP polymorphism in Nigerian sheep.
Also, we examined the LD among the 19 SNPs detected with others from previous studies using Lewontin’s D’ (|D’|) values (). The majority of the SNPs showed negative LD ranging from −0.018 to −0.188, except c.711 G > C which showed strong positive LD values of 0.135 and 0.095 with some SNPs.
Table 2. Linkage Disequilibrium (LD) of nineteen (19) PRNP polymorphisms in Nigerian sheep.
Haplotype frequency of the 14 PRNP nonsynonymous SNPs were further examined as shown in . Based on the haplotype analysis, we found eight (8) major haplotypes with the haplotype QMAGGHRRQQYNNS having the highest frequency (58.0%) followed by QMAGGRRRQQYNNS (13.7%) and QMAGGHRRQQDKKS (1.5%).
Table 3. Haplotype frequencies of fourteen (14) non-synonymous single nucleotide polymorphism of PRNP gene in Nigerian sheep.
Estimation of potential scrapie vulnerability in Nigerian sheep
To estimate potential scrapie vulnerability in Nigerian sheep, we compared the genetic distribution of scrapie-related SNPs (R154H and S240P) between Nigerian sheep and scrapie-affected sheep in other countries. Previous studies related to SNPs of ovine PRNP gene were selected to estimate the susceptibility in Nigerian sheep [Citation30,Citation40].
There was a significant difference (P<0.05) in allele frequencies of scrapie-affected Italian sheep and healthy Nigerian sheep counterpart at PRNP codon 143 (). However, no significant (P = 0.0064) difference was recorded between the allele frequencies of scrapie-affected Chinese sheep and Nigerian sheep at codon 143 ().
Figure 1. Comparisons of the allele frequencies of PRNP codons 154 and 240 in Central and Southern Italy, Chinese and Nigerian sheep. (a) Comparisons of the allele frequency of the PRNP codon 154 between Central and Southern Italy, Chinese sheep and Nigerian sheep [Citation30,Citation63] (b) Comparisons of the allele frequency of PRNP codon 240 between Chinese and Nigerian sheep [Citation30].
![Figure 1. Comparisons of the allele frequencies of PRNP codons 154 and 240 in Central and Southern Italy, Chinese and Nigerian sheep. (a) Comparisons of the allele frequency of the PRNP codon 154 between Central and Southern Italy, Chinese sheep and Nigerian sheep [Citation30,Citation63] (b) Comparisons of the allele frequency of PRNP codon 240 between Chinese and Nigerian sheep [Citation30].](/cms/asset/ca24d4e0-cdb2-46d2-82c2-ed88680e0c9a/kprn_a_2186767_f0001_oc.jpg)
Moreover, the allele frequencies of scrapie-affected Chinese and Nigerian sheep had no significant (P = 0.152) difference at codon 154 ().
Comparison of haplotype and genotype frequencies at PRNP codons 143, 154 and 171 between scrapie-affected and healthy sheep
The ovine PRNP haplotypes of codons 143,154 and 171 in Nigerian sheep were used for comparison with other previously published works on PRNP in sheep. Moreover, the PRNP haplotypes were compared with those previously reported in two countries i.e. Italian and Spanish sheep [Citation7,Citation40]. In the considered countries, the ARR and AHQ haplotypes were significantly different between the scrapie-affected and healthy sheep (). In Italian and Spanish sheep, ARQ haplotype was present but was not in Nigerian sheep. The genotypes ARR/ARR and ARQ/ARQ were significantly different between the scrapie-affected and healthy sheep.
Table 4. Distributions of haplotype and genotype frequencies at PRNP codons 143, 154 and 171 between scrapie affected and healthy sheep.
Assessment of nonsynonymous SNPs of the PRNP gene in Nigerian sheep
PolyPhen-2 is an online tool used to predict the outcome of an amino acid substitution caused by nonsynonymous SNPs on the structure and function of proteins [Citation64]. Based on our polymorphism results, the effects of 14 non-synonymous SNPs identified were assessed via PolyPhen-2. Three different predictions were observed for the 14 non-synonymous SNPs which include benign: T112M (0.000), S127G (0.000), S129G (0.000), H143R (0.241), H171Q (0.000), N176K (0.002), probably damaging: A116P (1.000), R151G (1.000), R154H (0.998), Y172D (1.000), and possibly damaging: Q101R (0.621), S240P (0.827) as shown in . We predicted the biological impact of the nonsynonymous SNPs identified with PROVEAN [Citation65]. Interestingly, the nonsynonymous SNPs of the PRNP gene identified in this study were predicted as ‘neutral’ (). Finally, we explored the amyloid propensity of ovine prion protein using the alleles of nonsynonymous SNPs. Based on this, we analysed the prion protein of these alleles and the HYNN haplotype was estimated with 0.00 values by AMYCO. Additionally, we evaluated the prion protein of Nigerian sheep. We classified amino acid sequences of PRNP in Nigerian sheep into three haplotypes (HYKK, HDKK and RYNN) considering the alleles of the 14 nonsynonymous SNPs. Based on AMYCO score, Haplotypes HYKK and HDKK when evaluated had a value of 0.00, while RYNN haplotype gave a value of 0.08 ().
Figure 2. Prediction of amyloid propensity of sheep prion protein according to nonsynonymous SNPs. AMYCO predicted amyloid propensity as values from 0.0 to 1.0. The AMYCO scores<0.45 and>0.78 indicated low and high aggregation propensities of the protein, respectively. ‘HYNN’ indicates haplotype of serine allele at the codon 139, serine allele at the codon 146, histidine allele at the codon 154, and Isoleucine allele at the codon 193. ‘HYKK’ indicates haplotype of Arginine allele at the codon 139, asparagine allele at the codon 146, arginine allele at the codon 154, and threonine allele at the codon 193. ‘HDKK’ indicates haplotype of arginine allele at the codon 139, asparagine allele at the codon 146, histidine allele at the codon 154, and threonine allele at the codon 193. ‘RYNN’ indicates haplotype of arginine allele at the codon 139, asparagine allele at the codon 146, arginine allele at the codon 154, and isoleucine allele at the codon 193.
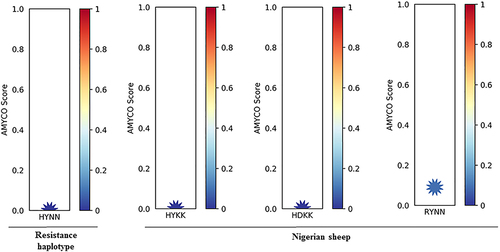
Table 5. Measurement of the effect of amino-acid substitutions of PRNP nonsynonymous SNPs in Nigerian sheep.
Discussion
Scrapie is a fatal disease in sheep but the distribution of the PrP genotypes could serve as a means to prevent subsequent reoccurrence in breeding programme. This study analysed the nucleotide sequence of the PRNP gene in 126 Nigerian sheep and revealed 19 SNPs of which 14 were non-synonymous. Interestingly, one novel SNP (T718C) was identified.
The polymorphism of the PRNP gene at codons 136 [Alanine (A) or Valine (V)], 154 [arginine (A) or histidine (H)], and 171 [R, H or glutamine (G)] are highly related to vulnerability or resistance to scrapie in sheep [Citation66–68]. The allelic variants (VRQ and ARQ) of PRNP at codons 136, 154 and 171 have been reported to correlate with high resistance to scrapie [Citation69,Citation70]. Moreover, the ARR allele was associated with low resistance to scrapie [Citation71]. In contrast, the VRQ allele at the same codon had low survivability after exposure to scrapie [Citation72]. Based on this, there is a need to develop breeding programs that will increase the frequency of the ARR allele [Citation73,Citation74]. It is believed that increasing such scrapie-resistant genotypes will enhance scrapie control.
In this present study, the non-synonymous substitutions i.e. Q101R, T112M, A116P, S127G, S129G, H143R, R151G, R154H, H171Q, Y172D, N176K, and S240P were identified (). The mutation at codon 127 was highly polymorphic similar to the trend detected in scrapie susceptibility in three native Ethiopian sheep breeds [Citation46]. It has been reported that amino acids at codons 126 and 127 are found in highly conserved glycine-rich motif.
GAVVG126G127LGGYMLG could reduce development of prion disease via blockage of amyloid fibril formation [Citation13]. In a previous, polymorphism at codon 127 was found to play a crucial role in the normal cellular function of PRNP [Citation75]. Moreover, the genotypes at codon 171Q/K are also related to scrapie susceptibility [Citation76].
The present study revealed that Nigerian sheep have different polymorphic sites mainly discovered at codons 112, 127, 129, 154, 171 and 176. Variations of PRNP at codons 112 and 129 have not been reported in sheep and their functions of codons 112 and 129 have not been established. However, we only detected polymorphism at codons 154 and 171, which are susceptible to classical scrapie in sheep.
Furthermore, PolyPhen-2 and PROVEAN were used to predict the impacts of 14 non-synonymous SNPs identified in this study. Using PolyPhen-2, three different predictions were observed for the 14 non-synonymous SNPs which include benign: T112M (0.000), S127G (0.000), S129G (0.000), H143R (0.241), H171Q (0.000), N176K (0.002), probably damaging: A116P (1.000), R151G (1.000), R154H (0.998), Y172D (1.000), and possibly damaging: Q101R (0.621), S240P (0.827). Although, all the 14 non-synonymous SNPs identified for the PRNP gene in Nigerian sheep populations were ‘(neutral or non-deleterious)’ using PROVEAN. The discrepancies in the prediction outcomes of the two software might be due to variations in algorithms that reproduce the effect on the position of the protein [Citation64,Citation77] and difference in their mode of analyses [Citation78]. We investigated the amyloid propensity of sheep prion protein based on the alleles of nonsynonymous SNPs of the PRNP gene in sheep. We found out that the amyloid formation of PRNP in two of the haplotypes (HYKK and HDKK) was similar to the resistance haplotype (HYNN) to prion diseases in this study. However, one of the haplotypes (RYNN) had a higher degree of amyloid formation value than the resistance haplotype ().
Conclusion
In conclusion, the polymorphism identified in this study show that sheep in Nigeria are susceptible to scrapie because of the variations detected at codons 154 and 171. The information obtained about the variation in allelic frequencies of PRNP gene in Nigerian sheep could assist into designing valuable scrapie-resistance breeding projects especially for sheep in a tropical region like Nigeria, thereby reducing disease outbreaks caused by scrapie and the subsequent increase in cost of production.
Author contributions:
Conceptualization, A.C.A., S.F.B., and A.M.A.; methodology, A.C.A., S.F.B., A.M.A., L.M.N., A.B.O, L.D.R., T.T.Y., and N.A.; software, A.C.A., and S.F.B.; validation, S.F.B., A.M.A., L.M.N., A.B.O., L.D.R., P.M.D., and N.A.; formal analysis, A.C.A., and S.F.B.; investigation, A.C.A., S.F.B., S.C.O., and M.P.H.; resources, A.C.A., A.M.A, A.B.O., N.A., L.D.R., S.L.P., M.M.M., H.S., A.I.M., O.J.S., L.M.N., S.C.O., G.F.M., M.P.H., R.K.B., J.I., H.L.A., and P.M.D.; data curation, A.C.A., S.F.B., and T.T.Y.; writing – original draft preparation, S.F.B., and A.C.A.; writing – review and editing, A.E.S., A.M.A., A.B.O., N.A., L.M.N., S.L.P., M.M.M., H.S., R.K.B., H.L.A., A.I.M., O.J.S., L.D.R., S.C.O., G.F.M., M.P.H., J.I., S.K., M.H.Y., and P.M.D.; visualization, A.C.A., S.F.B., A.M.A., S.K., P.M.D., and A.E.S; supervision, A.C.A.; project administration, A.C.A.; funding acquisition, A.C.A. All authors have read and agreed to the published version of the manuscript.
Institutional review board statement
All experimental procedures in this study were in accordance with Research Guidelines for the Institutional Review Board of Kunming Institute of Zoology, Chinese Academy of Sciences (SMKX-20160524-119) and the Central Abattoir, Ibadan, Ministry of Agriculture and Rural Development, Oyo State, Nigeria. Various sheep were transported from farms in drier agro-climate zones in Nigeria to the Central Abattoir in Ibadan, Nigeria. Hence, all sheep samples in this study were collected at the Central Abattoir, Ibadan.
Informed consent statement:
Not applicable.
Supplemental Material
Download Zip (3.4 MB)Acknowledgments
We appreciate the important contributions of the Central Abattoir, Ibadan and the Ministry of Agriculture and Rural Development, Oyo State, in Nigeria, who made possible the collection of sheep samples for this study. We recognize all of those who assisted in the success of this study.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
Accession no. MZ463500 – MZ463625 https://www.ncbi.nlm.nih.gov/
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/19336896.2023.2186767.
Additional information
Funding
References
- Glatzel M, Aguzzi A. The shifting biology of prions. Brain Res Rev. 2001;36:241–248.
- Tatzelt J. Prion proteins. Top Curr Chem 2011;305: ix–x.
- Ascari LM, Rocha SC, Gonçalves PB, et al. Challenges and advances in antemortem diagnosis of human transmissible spongiform encephalopathies. Front Bioeng Biotechnol. 2020;8:1–22.
- Slapšak U, Salzano G, Ilc G, et al. Unique structural features of mule deer prion protein provide insights into chronic wasting disease. ACS Omega. 2019;4:19913–19924.
- Goldmann W. PrP genetics in ruminant transmissible spongiform encephalopathies. Vet Res. 2008;39:30.
- Langeveld JPM, Pirisinu L, Jacobs JG, et al. Four types of scrapie in goats differentiated from each other and bovine spongiform encephalopathy by biochemical methods. Vet Res. 2019;50. DOI:10.1186/s13567-019-0718-z.
- Acín C, Martín-Burriel I, Goldmann W, et al. Prion protein gene polymorphisms in healthy and scrapie-affected Spanish sheep. J Gen Virol. 2004;85:2103–2110.
- Novák M, Vrtiak OJ, Mikula I, et al. Ovine scrapie: priorities and importance. Folia Microbiol (Praha). 2000;45:475–483.
- Dassanayake RP, Madsen-Bouterse SA, Truscott TC, et al. Classical scrapie prions are associated with peripheral blood monocytes and T-lymphocytes from naturally infected sheep. BMC Vet Res. 2016;12. DOI:10.1186/s12917-016-0651-6
- Dassanayake RP, Schneider DA, Truscott TC, et al. Classical scrapie prions in ovine blood are associated with B lymphocytes and platelet-rich plasma. BMC Vet Res. 2011;7:75.
- Arsac JN, Bétemps D, Morignat E, et al. Transmissibility of atypical scrapie in ovine transgenic mice: major effects of host prion protein expression and donor prion genotype. PLoS ONE. 2009;4:e7300.
- Goldmann W, Ryan K, Stewart P, et al. Caprine prion gene polymorphisms are associated with decreased incidence of classical scrapie in goat herds in the United Kingdom. Vet Res. 2011;42:42.
- Zhang J, Zhang Y. Molecular dynamics studies on 3D structures of the hydrophobic region PrP(109-136). Acta Biochim Biophys Sin (Shanghai). 2013;45:509–519.
- Baral PK, Yin J, Aguzzi A, et al. Transition of the prion protein from a structured cellular form (PrPC) to the infectious scrapie agent (PrPsc). Protein Sci. 2019;28:2055–2063.
- Requena JR. The protean prion protein. PLoS Biol. 2020;18:1–8.
- Sanz-Hernández M, Barritt JD, Sobek J, et al. Mechanism of misfolding of the human prion protein revealed by a pathological mutation. Proc Natl Acad Sci U S A. 2021;118. DOI:10.1073/pnas.2019631118
- Gelasakis AI, Boukouvala E, Babetsa M, et al. Polymorphisms of codons 110, 146, 211 and 222 at the goat prnp locus and their association with scrapie in greece. Animals. 2021;11:2340.
- Greenlee JJ. Review: update on classical and atypical scrapie in sheep and goats. Vet Pathol. 2019;56:6–16.
- Cassmann ED, Greenlee JJ. Pathogenesis, detection, and control of scrapie in sheep. Am J Vet Res. 2020;81:600–614.
- Yang S, Thackray AM, Hopkins L, et al. Polymorphisms at amino acid residues 141 and 154 influence conformational variation in ovine PrP. BioMed Res Int. 2014;2014:1–14.
- Andrade CP, Neto JDB, Driemeier D. Identification of single nucleotide polymorphisms in the prion protein gene in Santa Ines and Dorset sheep. Pesqui Vet Bras. 2018;38:624–628.
- Acín C, Bolea R, Monzón M, et al. Classical and atypical scrapie in sheep and goats: review on the etiology, genetic factors, pathogenesis, diagnosis, and control measures of both diseases. Animals. 2021;11:1–20.
- Cassmann ED, Mammadova N, Jo Moore S, et al. Transmission of the atypical/Nor98 scrapie agent to Suffolk sheep with VRQ/ARQ, ARQ/ARQ, and ARQ/ARR genotypes. PLoS One. 2021;16:1–8. DOI:10.1371/journal.pone.0246503.
- Leal JS, de Andrade CP, Correa GLF, et al. Classical scrapie diagnosis in ARR/ARR sheep in Brazil. Acta Sci Vet. 2015;43:7.
- Boareki M, Schenkel F, Kennedy D, et al. Prediction of genetic resistance for scrapie in ungenotyped sheep using a linear animal model. Genes (Basel). 2021;12:1432.
- Harrathi C, Fernández-Borges N, Eraña H, et al. Insights into the bidirectional properties of the sheep–deer prion transmission barrier. Mol Neurobiol. 2019;56:5287–5303.
- Li J, Zhang S, Erdenee S, et al. Nucleotide variants in prion-related protein (testis-specific) gene (PRNT) and effects on Chinese and Mongolian sheep phenotypes. Prion. 2018;12:185–196.
- Li J, Erdenee S, Zhang S, et al. Genetic effects of PRNP gene insertion/deletion (indel) on phenotypic traits in sheep. Prion. 2018;12:42–53.
- Lan Z, Wang ZL, Liu Y, et al. Prion protein gene (PRNP) polymorphisms in Xinjiang local sheep breeds in China. Arch Virol. 2006;151:2095–2101.
- Xu L, Zhang Z, Zhou X, et al. Molecular cloning and polymorphism analysis of the prion protein gene in Tan sheep of Ningxia, China. Gene. 2011;485:102–105.
- Vitezica ZG, Beltran de Heredia I, Ugarte E. Short communication: analysis of association between the prion protein (PRNP) locus and milk traits in Latxa dairy sheep. J Dairy Sci. 2013;96:6079–6083.
- Gerlinger C, Verrier E, Goritschnig S. International Congress on the Breeding of Sheep and Goats. Genet Resour. 2021;2:1–114.
- Koynarski T. Prp gene polymorphism and its influence on some productive traits of sheep breeds reared in Bulgaria. Bulg J Vet Med. 2020;23:170–177.
- González L, Pitarch JL, Martin S, et al. Influence of polymorphisms in the prion protein gene on the pathogenesis and neuropathological phenotype of sheep scrapie after oral infection. J Comput Pathol. 2014;150:57–70.
- Álvarez L, Gutiérrez-Gil B, San Primitivo F, et al. Influence of prion protein genotypes on milk production traits in Spanish churra sheep. J Dairy Sci. 2006;89:1784–1791.
- Argyriadou A, Gelasakis AI, Banos G, et al. Genetic improvement of indigenous Greek sheep and goat breeds. J Hell Vet Med Soc. 2020;71:71.
- O’doherty E, Aherne M, Ennis S, et al. Prion protein gene polymorphisms in pedigree sheep in Ireland. Res Vet Sci. 2001;70:51–56.
- de Andrade CP, de Almeida LL, de Castro LA, et al. Development of a real-time polymerase chain reaction assay for single nucleotide polymorphism genotyping codons 136, 154, and 171 of the prnp gene and application to Brazilian sheep herds. J Vet Diagnostic Investig. 2013;25:120–124.
- Martin Palomino P, Gomez Calle L, Serejo Proença J, et al. Genotyping of prion protein in black merino sheep from the Iberian Peninsula. Small Rumin Res. 2019;173:36–41.
- Curcio L, Sebastiani C, Ceccobelli S, et al. PRNP polymorphisms in four Italian sheep breeds. Livest Sci. 2015;181:38–42.
- Meydan H, Pehlivan E, Özkan MM, et al. Prion protein gene polymorphisms in Turkish native goat breeds. J Genet. 2017;96:299–305.
- Alsayed O, Erkunt Alak S, C Ü. Analysis of prion protein coding gene polymorphisms in Palestinian native sheep breeds. Ankara Univ Vet Fak Derg. 2019;66. DOI:10.33988/auvfd.437314
- Han CX, Liu HX, Lu YX, et al. Polymorphism of prion protein gene in sheep of Inner Mongolian, China. Virus Genes. 2011;42:153–155.
- Zhao CL, Wu R, Liu L, et al. Ovine prion protein genotype frequencies in northwestern China. Genet Mol Res. 2012;11:1671–1681.
- Djaout A, Chiappini B, Gaouar SBS, et al. Biodiversity and selection for scrapie resistance in sheep: genetic polymorphism in eight breeds of Algeria. J Genet. 2018;97:453–461.
- Teferedegn EY, Yaman Y, Un C. Five novel PRNP gene polymorphisms and their potential effect on Scrapie susceptibility in three native Ethiopian sheep breeds. BMC Vet Res. 2020;16. DOI:10.1186/s12917-020-02336-0
- Serrano C, Martín-Burriel I, Lyahyai J, et al. Polymorphisms of the PRNP gene in Moroccan sheep breeds. Vet Rec. 2007;161:524–525.
- Traoré A, Royo LJ, Kaboré A, et al. Prion protein gene polymorphism in four West African sheep populations. Trop Anim Health Prod. 2012;44:1469–1472.
- Lombin LH Report on the internet [Internet]. 2007; Available from: http://www.africanagricultureblog.com/2007/12/nigeria-has-16-million-cattle.html.
- Yunusa AJ, Salako AE, Oladejo OA. Morphometric characterization of Nigerian indigenous sheep using multifactorial discriminant analysis. Int J Biodiver Conserv. 2013;5:661–665.
- Yakubu A. Multivariate analysis of morphostructural characteristics in Nigerian indigenous sheep. Ital J Anim Sci. 2011;10:83–86.
- Pona HT, Duan X, Ayantobo O, et al. Environmental health situation in Nigeria: current status and future needs. Heliyon. 2021;7:e06330.
- Agaviezor B, Peters S, Adefenwa M, et al. Morphological and microsatellite DNA diversity of Nigerian indigenous sheep. J Anim Sci Biotechnol. 2012;3:38.
- Adebambo O, Williams JL, Blott S, et al. Genetic relationships between Native Sheep breeds in Nigeria based on microsatellite DNA polymorphisms. Anim Genet Resour Inf. 2004;34:27–39.
- Sheriff O, Alemayehu K, González-Redondo P. Genetic diversity studies using microsatellite markers and their contribution in supporting sustainable sheep breeding programs: a review. Cogent Food Agric. 2018;4:1459062.
- Okpeku M, Ogah DM, Adeleke MA. A review of challenges to genetic improvement of indigenous livestock for improved food production in Nigeria. Afr J Food Agric Nutr Dev. 2019;19:13959–13978.
- Iroanya G, Osaiyuwu O, Emmanuel H, et al. Genetic polymorphism of myostatin (MSTN) in Nigerian sheep breeds. 2021;6:64–73. DOI:10.31248/JASVM2021.257.
- Osaiyuwu O, Salako E. Genetic structure of indigenous sheep breeds in Nigeria based on electrophoretic polymorphous systems of transferrin and haemoglobin. Afr J Biotechnol. 2018;17:380–388.
- Nwankiti OO, Ikeh EI, Arowolo OA, et al. A targeted survey for scrapie in Jos Plateau State, Nigeria. J Vet Med. 2013;2013:1–5.
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual (3-volume set). Cold Spring Harbor Labpratory Press; 2001.
- Rozas J, Ferrer-Mata A, Sanchez-DelBarrio JC, et al. DnaSP 6: dNA sequence polymorphism analysis of large data sets. Mol Biol Evol. 2017;34:3299–3302.
- Iglesias V, Conchillo-Sole O, Batlle C, et al. AMYCO: evaluation of mutational impact on prion-like proteins aggregation propensity. BMC Bioinf. 2019;20:20.
- Torricelli M, Sebastiani C, Ciullo M, et al. PRNP polymorphisms in eight local goat populations/breeds from Central and Southern Italy. Anim an Open Access J from MDPI. 2021;11:333.
- Goldmann W, Hunter N, Smith G, et al. PrP genotype and agent effects in scrapie: change in allelic interaction with different isolates of agent in sheep, a natural host of scrapie. J Gen Virol. 1994;75:989–995.
- Choi Y, Sims GE, Murphy S, et al. Predicting the functional effect of amino acid substitutions and indels. PLoS ONE. 2012;7:e46688.
- Tongue SC, Wilesmith JW, Cook CJ. Frequencies of prion protein (PrP) genotypes and distribution of ages in 15 scrapie-affected flocks in Great Britain. Vet Rec. 2004;154:9–16.
- Vaccari G, Petraroli R, Agrimi U, et al. PrP genotype in Sarda breed sheep and its relevance to scrapie. Arch Virol. 2001;146:2029–2037.
- Baylis M, Goldmann W. The genetics of scrapie in sheep and goats. Curr Mol Med. 2005;4:385–396.
- Clouscard C, Beaudry P, Elsen JM, et al. Different allelic effects of the codons 136 and 171 of the prion protein gene in sheep with natural scrapie. J Gen Virol. 1995;76:2097–2101.
- Baylis M, Goldmann W, Houston F, et al. Scrapie epidemic in a fully PrP-genotyped sheep flock. J Gen Virol. 2002;83:2907–2914.
- Kutzer T, Pfeiffer I, Brenig B. Identification of new allelic variants in the ovine prion protein (PrP) gene. J Anim Breed Genet. 2002;119:119.
- Elsen JM, Amigues Y, Schelcher F, et al. Genetic susceptibility and transmission factors in scrapie: detailed analysis of an epidemic in a closed flock of Romanov. Arch Virol. 1999;144:431–445.
- Ponz R, Tejedor MT, Monteagudo LV, et al. Scrapie resistance alleles are not associated with lower prolificity in Rasa Aragonesa sheep. Res Vet Sci. 2006;81:37–39.
- Un C, Oztabak K, Ozdemir N, et al. Genotyping of PrP gene in native Turkish sheep breeds. Small Rumin Res. 2008;74:260–264.
- Vitale M, Migliore S, Tilahun B, et al. Two novel amino acid substitutions in highly conserved regions of prion protein (PrP) and a high frequency of a scrapie protective variant in native Ethiopian goats. BMC Vet Res. 2019;15. DOI:10.1186/s12917-019-1870-4
- Cassmann ED, Moore SJ, Smith JD, et al. Sheep with the Homozygous Lysine-171 prion protein genotype are resistant to classical scrapie after experimental oronasal inoculation. Vet Pathol. 2019;56:409–417.
- Walters-Sen LC, Hashimoto S, Thrush DL, et al. Variability in pathogenicity prediction programs: impact on clinical diagnostics. Mol Genet Genomic Med. 2015;3:99–110.
- Kim SK, Kim YC, Won SY, et al. Potential scrapie-associated polymorphisms of the prion protein gene (PRNP) in Korean native black goats. Sci Rep. 2019;9:1–10. DOI:10.1038/s41598-019-51621-y.
