ABSTRACT
Domestication of orphan crops could be explored by editing their genomes. Genome editing has a lot of promise for enhancing agricultural output, and there is a lot of interest in furthering breeding in orphan crops, which are sometimes plagued with unwanted traits that resemble wild cousins. Consequently, applying model crop knowledge to orphan crops allows for the rapid generation of targeted allelic diversity and innovative breeding germplasm. We explain how plant breeders could employ genome editing as a novel platform to accelerate the domestication of semi-domesticated or wild plants, resulting in a more diversified base for future food and fodder supplies. This review emphasizes both the practicality of the strategy and the need to invest in research that advances our understanding of plant genomes, genes, and cellular systems. Planting more of these abandoned orphan crops could help alleviate food scarcities in the challenge of future climate crises.
1. Introduction
Orphan crops are also known as ‘underutilized,’Citation1, ‘minor,’Citation2, ‘neglected,’Citation3,Citation4 ‘promising’ (for emerging markets or because of previously unrecognized valuable traits), ‘niche’ (of marginal importance in production systems and economies) and/or ‘traditional’ (used for centuries or even millennia) crops.Citation5 These crops show many important characteristic features and are very much fitting to be grown as cash crops. Ironically, these crops are less familiar globally, primarily due to less attention paid by researchers, leading to inadequate or total lack of genetic and genomic resources. Despite the negligence in research and investment, they still have the potential to tackle multiple UN Sustainable Development Goals (such as zero hunger) in the low-income nations of Africa,Citation3 Latin America, and Asia,Citation3 and also in the growing western consumers interested in new healthier foods.Citation6
Most orphan crops are resilient and can grow on extreme soil and climatic conditions as they possess relevant alleles and mechanisms to combat stress conditions,Citation7 potentially lost from major crops.Citation8 In due course of time, it has been recognized that orphan crops possess resilience traitsCitation9 and can be used to improve major crops as well as play an essential role in enhancing the sustainability of food systemsCitation6,Citation10,Citation11 which in turn has resulted in the launch of advanced research and development initiatives.Citation4 It has also been seen that orphan crops possess other traits of importance that include nutrition,Citation6 biofuel,Citation12 medicinal value,Citation13 cosmetics,Citation14 and feed/fodder.Citation15
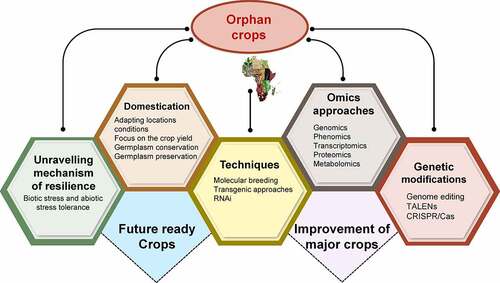
The research and breeding efforts were majorly focused on some of the few crops on which the world’s food supply relies, like rice, wheat, maize, soy, and potato. For instance, by identifying the wild rice allotetraploids, the important wild sources of de novo domestication can be used by tissue culture, transfronation and genome editing systems to domesticate the wild rice races.Citation16 In addition to these crops variety of orphan crops was also grown by small farmers.Citation17,Citation18 Orphan crops are a set of nutritious, tasty, and well-adapted species but, due to wild characteristics, are often unsuited to intensive agriculture.Citation19,Citation20 Orphan crops possess many characteristics and can be a lifesaver to the millions who go to bed hungry every night.Citation20 Besides, orphan crops served as resource material for agricultural research to further increase the stress tolerance of major food crops and also brought diversity to agriculture.Citation20 In addition, it also favored shifting food habits of modern agriculture. It has been proven that most orphan crops such as buckwheat (Fagopyrum esculentum), cassava, banana, and quinoa are not only important for food and feed but also hold great potential for industrial applications, effective management and have contributed toward poverty reduction and global food security.Citation21,Citation22 Conventional breeding for quality enhancement in orphan crops is challenging, while genetic manipulation via guided nucleases has proven an ideal platform for improving orphan cropsCitation23–25 (). This robust domestication was used to tackle changing climate scenarios and growing food security issues. Therefore, the current review aims to emphasize the practicality of the strategy and the need to invest in exploring the genomes, proteomics, and other omics studies to unravel their potential as frontline crops. In our investigation and exploration, we have discussed the updated account on orphan crops and their potential to act as buffering agents for securing nutrition and food security at global level. More importantly, we have detailed their social, ethical and legal point of consideration to have a global overview to make sure their role as multidimensional crops for future.
2. Orphan Crops: A Rich Repository Crop for Food and Nutritional Security
Orphan crops (crops for the future) are cultivated in very limited regions at global level. The reason for this low cultivation may be due to less research involved in to unravell their resilient characteristics.Citation4,Citation6,Citation26,Citation27 The worldwide economic value of orphan crops is limited but is extremely important at local levels, especially in developing countries.Citation9 These crops mainly include cereals, pseudo cereals, legumes, and root crops, contributing to food and nutritional needs worldwide. These crops possess many properties like being resilient to biotic and abiotic stresses, high nutritional and medicinal value, and high photosynthetic efficiency. Examples of orphan cereals that utilize highly efficient C4 photosynthesis pathways include foxtail millet, pearl millet, broomcorn millet, barnyard millet, fonio millet, adlay, finger millet, and tef.Citation28–30 C4 photosynthesis promotes the efficient use of nitrogen and water by reducing photorespiration to a minimum in hot and arid climates compared to C3 photosynthesis. Wild relatives also adopt the C4 pathway.Citation31 Some of the orphan crops and their characteristic properties are given in .
Table 1. Showing the detail of major orphan crops and their characteristic traits considered for domestication process.
Orphan crops, as already discussed, possess many important features and can be a lifesaver to the millions of hungry humans on global scale. As stress-tolerant in nature, these crops can grow under unpredictable changing climates and ensure yield when major crop varieties fail, thus ensuring food security for small farmers. Besides, orphan crops served as resource material for agricultural research to further increase the stress tolerance of major food crops and also brought diversity to agriculture. In addition, these crops also favored shifting food habits of modern agriculture.Citation31,Citation53 It has been seen that orphan crops and their wild relatives possess many beneficial traits compared to major crops, which can be used to overcome different problems of modern agriculture and help make the practice more sustainable. Since most of these crops were well suited to grow in poor and marginal lands, there was little need for irrigation and fertilizers to boost their productivity. Coupling these crops with various sustainable agricultural techniques such as crop rotation and intercropping have been proven to be the best approaches to maintain a high yield of industrial agriculture while having a minimal ecological impact.Citation54 Intercropping of some orphan crops like quinoa with beans, corn, and tarwi improved its yield and helped in pest control.Citation55 Crop rotation of broomcorn millet with other crops prevented diseases/posts, helped in weed control, and helped maintain soil moisture.Citation56
Despite possessing these properties, some orphan crops are still not domesticated and possess a lot of potential in terms of quality and quantity of yield. Domestication of various crops through past approaches can lead to the faster improvement of orphan crops.Citation57 In addition to these approaches latest developing techniques like high-throughput sequencing and genome editing can play an important role in the genetic improvement and manipulation of these minor crops. These crops possess many properties like being resilient to biotic and abiotic stresses, high nutritional and medicinal value, and high photosynthetic efficiency. As orphan crops are already comparatively more stress tolerant and nutritious, domestication efforts must mainly concentrate on boosting the yield.Citation49,Citation57 Some gene-controlling characteristics often conserved include grain size, weight, and height, and they can be directly targeted using advanced genome editing techniques like clustered regularly interspaced short palindromic repeats (CRISPR/Cas9) systems (). Sequencing the genome of these crops leads to large-scale improvement of these crops and makes them available publicly. In addition, this also served as a solid foundation for the modifications and development of these crops via various techniques like molecular breeding and genome editing.Citation49
3. Genomic Information on Orphan Crops
The molecular breeding has been revolutionized due to the advance in genomics and genome editing technologies.Citation58–61 The valuable information of genomic data gained from high-throughput sequencing and computational analysis can be accomplished to identify desirable traits incorporated in wild relatives of crop plants.Citation59,Citation61 Many initiatives have been taken for advanced research and development of orphan crops. Different institutions have taken the step for the detailed analysis of some crops; 101 orphan crops have been selected by African orphan Crops Consortium (AOCC) for detailed studies and whole-genome sequencing.Citation62 Next-generation sequencing, in which whole-genome sequencing is also available for many crops and their wild relatives, has proven an efficient way of facilitating the domestication of genes. Thus, crop improvement has been “democratized.”Citation63 The availability of whole-genome sequencing provides information about ortholog identities and selective sweeps and is an essential requirement for both source and recipients of allelic variance. In addition, this application of high-quality sequencing provides necessary information about three features of domestication, which are genomic structural variation, transposable elements, and gene as well as whole-genome duplications.Citation64 Studies have shown that genomic structural variation concerns some important agricultural traits, copy number variation like variation in vernalization and flowering time in wheatCitation65 and barley freezing tolerance.Citation66 At high frequencies, transposable elements are present all over the whole genome.Citation67 Whole-genome duplication has also proven significant by revealing almost 200 crops and over 2000 wild species.Citation68 In recent years, significant achievements in genome sequencing have been achieved in the grass family, and reference genomes of at least 11 cereals have been published. The orphan cereal which was first sequenced was foxtail millet.Citation69 Sequencing of orphan legumes has also been done, and at least the genome of 16 orphan legumes have been sequenced; the orphan legume whose genome was first sequenced was pigeon pea.Citation26 The genome of other orphan crops, which has also been sequenced, includes pseudocereals amaranth,Citation70 buckwheat, and quinoaCitation71 and the root crops cassava,Citation72 sweet potato,Citation73 and yams.Citation74 Besides genome sequencing, transcript profiling, epigenetics studies, and the use of metabolomics also provide considerable insight into domestication.Citation57,Citation64,Citation75
4. Omics Studies and Their Utilization for Desired Genome Editing
Genome sequencing technologies have undergone rapid evolution, which enables the explosion of large-scale data at each level of information that is from gene sequence, transcriptome, proteome, epigenome as well as metabolite patterns that usually determine the variability in cellular networks and functions at the systems level.Citation75–79 The information gained by the multi-omics approach can be integrated to pinpoint the molecular determinants and provide a platform to improve crop yield and quality traits in orphan crops.Citation80
The genome of orphan crops was well annotated by whole-genome sequencing projects that were often coupled with generating respective transcriptomes. The transcriptomes that were mainly preferred for annotation were taken from the same species as was done for the African eggplant,Citation81 wild mustard (Brassica juncea),Citation82 and tef (Eragrostis tef)Citation28; however, some other cases were also seen where there was lack of resources, existing transcriptome of a close relative or well-annotated transcriptome of a model crop was used as seen in finger millet, the genome was annotated by using data from maize.Citation30 Some of the other transcriptomes of these crops have also been generated in response to specific biological questions, and RNA sequencing (RNA-seq) has become the method of choice.Citation83 Researchers identified at least 2416 differentially expressed genes while profiling for response against salt stress in quinoa (Chenopodium quinoa).Citation84 Transcription analysis was done in jute-mallow to identify drought stress-related genes.Citation73 Before the introduction of next-generation sequencing (NGS), microarrays were the methods of choice for transcriptome analysis. They were applied in different orphan crops to detect expression profiles relevant to abiotic stress resilience. These crops of interest include buckwheat,Citation85 tef,Citation85 white lupine,Citation86 African nightshade (Solanum nigrum),Citation87 and wild mustard.Citation88 Epigenetics is a branch of science that deals with studying heritable gene regulation. The epigenetic changes do not involve DNA sequences but occur by the modifications caused by DNA methylation or post-translational modification of histone tails. These changes are believed to play an important role in gene expression and plant development under stress conditions (). The most important genomic approach, ChIP-chip (followed by microarray hybridization), was mainly used for analyzing epigenetic changes. Still, in recent years this approach was replaced by NGS technologies with ChIP-sequencing (ChIP followed by direct sequencing). Studies have shown that several epigenetic changes have contributed to crop domestication.
Figure 3. Displays the rich salient attributes of orphan crops fitting to human health and sustainable agricultural development for food and nutritional security.

Several epigenetic modifications have been found to contribute to crop domestication; for example, the colorless non-ripening locus in tomatoes is induced by silencing an epiallele of a SQUAMOSA promoter binding protein-like transcription factor.Citation89 Epialleles have also been linked to melon sex,Citation90 rice plant height,Citation91 cotton photoperiod sensitivity,Citation92 and oil palm somaclonal variation.Citation92,Citation93
In addition, genome-wide analyses of the methylomes of significant crop species indicate that the bulk of these alterations is substantially conserved within a species.Citation94 The use of metabolomics in the study of domestication is relatively new, but it is widely utilized in quantitative trait loci (QTL) investigationsCitation95; it is becoming a mainstay for understanding the genetics of quality traits in crops.Citation96,Citation97 Nonetheless, two studies address how domestication and agricultural development influence the metabolome. First, a comprehensive analysis of alterations in primary metabolism in farmed wheat and its progenitor species revealed that the domestication of emmer and durum wheat, respectively, is accompanied by changes in unsaturated fatty acid and amino acid content.Citation98 Secondly, a multi-omics analysis of fruit from several hundred tomato genotypes demonstrates how domestication has affected the metabolite composition of this fruit.Citation99
5. The Role of Genome Editing Technologies in the Orphan Crops Improvement
Genome editing has proven to be a powerful tool for crop improvement and functional genomics. Several gene-editing techniques have been widely used, including zinc-finger nucleases (ZFNs), transcription activator-like (TAL) effector nucleases (TALENs), and CRISPR/Cas9 and Cpf1. The aim of these techniques relies on the future improvement of plant sciences and the rapid remodeling of crops. Genome editing via CRISPR has become popular because of its versatility, potency, adequacy, and simplicity.Citation23,Citation100,Citation101
Improvement and domestication of crops have become possible due to the availability of genomic information and efficient genome editing toolsCitation102 for the domestication of these crops. Genome editing technologies played an important role in improving orphan crops by identifying important mechanisms and gene targets for domestication and making it possible to modify these crops in a targeted manner. CRISPR-mediated genome editing has proven more advantageous as it produces potentially ‘transgene-free’ crop varieties and provides a platform to create genetically modified varieties identical to conventionally bred crop varieties. For several cultivated species, the path of domestication has become evident due to the noble intervention of genetic and genomic analyses.Citation57 Editing plant genomes with extreme precision and accuracy are achieved by combining genome editing technologies, viz. CRISPR/Cas9. The CRISPR system is not entirely accepted as its acceptance is controversial, but despite this, it still holds great potential in improving crop varieties and improving farmer’s livelihood. DNA manipulation via CRISPR/Cas9 can be done in many ways like by causing random mutations (insertion or deletion) via non-homologous end joining for disruption of genes by the involvement of precise base editors to generate targeted point mutations or by whole-gene insertion employing the cell’s homology-directed repair pathway.Citation100 Multiplex approaches enabled researchers to edit multiple loci simultaneously and made it possible to incorporate multiple traits at once.Citation103
Domestication of orphan crops is a complicated process as it requires well-elucidated genome sequence that gives a clear understanding of paralogue structure and expression of genes, and a delivery system for genome editing. Among them, a transformation system is the simplest. Due to multigenic regulation of the same traits in orphan crops, their domestication is somehow limited.Citation104 Many domestication genes like that control flowering and fruit development, increase harvest index (more product per plant), facilitate harvesting by inhibiting abscission of fruits, or make the final product easier to store, chew, and digest have been identified in many crop species.Citation102 Studies have shown that mutations played an important role in altering the function of a few selected loci, known as domestication genes. Also, it has been revealed that many domestication traits possess a Mendelian inheritance pattern involving gain-of-function or loss-of-function mutations, which made it possible to modify/reconstruct these traits into suitable ones with the help of genome editing technologies, viz. CRISPR/Cas9 genome editing technology.Citation105 First, the applications of CRISPR/Cas9 were limited to creating only deletion. Still, modern variants of CRISPR-based genome editing technologies are more advantageous due to new modifications and can produce targeted insertions, exchange amino acids, and modulate gene expression. The variants generated by manipulating just a small number of loci were best suited to full agricultural exploitation. Also, they demonstrated the feasibilities for crop improvement that can be achieved through a combination of genomics and gene editing. Knockout may be inadequate in many cases and may need subtler alleles like altering promoter activity or protein structure. Production of alleles with new and valuable expression properties can be achieved by editing promoter segments.Citation106 Manipulating several genes with the connected combinatorial challenge of testing many variables leads to the acceleration of domestication. Hence, genome editing has been a powerful tool for domestication of wild plants and reuniting lost but desirable traits that include nutritional features or stress tolerance, yield potential, and other agronomically valuable characteristics.Citation107
With the CRISPR/Cas9 approach, some successful cases of orphan crop breeding and domestication of wild relatives have been reported recently. Scientists developed genomic resources and efficient transformation methods for orphan crop groundcherry (Physalis pruinosa) belonging to the Solanaceae family. They then mutated the genes orthologues to tomato domestication and improved genes using the CRISPR-Cas9 system to improve productivity-related traits.Citation108 The targeted genes include plant architecture, fruit size, and flower production. Studies have shown that manipulation via CRISPR/Cas9 in wild tomato (S. pimpinellifolium) involved editing six loci that were important for yield and productivity in present-day tomato crop lines.Citation109 These modifications proved very beneficial, like increased fruit size threefold, fruit number tenfold, and a two-fold increase in fruit lycopene accumulation compared to its wild parent. CRISPR/Cas9 system gained prominence, and it has also been seen that through this approach, several types of orphan crops can be modified and have been successfully used in foxtail millet,Citation110 green foxtail,Citation111 and cassava.Citation112
5.1. Prospective Applications of CRISPR System in Cassava Breeding: A Case Study
Cassava (M. esculenta), which is an important staple food, is grown worldwide and is a significant orphan crop not only for providing food security to tropical and subtropical regions but also being the predominant raw material for the starch industry and also provides up to 50% of the total intake calories for over 800 million people worldwide.Citation113 It possesses many resilient characteristics, like being tolerant to unpredicted drought, growing very well in poor soils, and can be harvested any time of the year. Also, it is advantageous as its tubers can be retained for up to two years in soil without rotting.Citation114 Despite possessing many properties, its yield is low, so many attempts are made to increase its yield, but compared to mainstream crops like rice, in cassava, there are only a few studies on the validity of the CRISPR technique. Recently many genome editing projects involving CRISPR/Cas9 have been completed to increase the yield of cassava, including disease resistance, herbicide tolerance, rapid flowering, and reduced cyanide content leaves and roots.Citation115 Disease-causing pathogens majorly cause the loss of yield in cassava. Up to 50% of the total yield loss is mainly caused by the African cassava mosaic virus (ACMV) and cassava brown streak disease (CBSD). So to overcome these problems, different strategies have been developed, viz. targeted mutation using Cas9/gRNA.Citation116 Gene editing of two isoforms of elF4E, nCBP-1 and nCBP-2, were done simultaneously, resulting in heritable delayed and suppressed CBSD aerial symptoms and reduced severity and frequency of storage root necrosis. Resistance against ACMV in cassava was achieved by knockout of the AC2 gene resulting in 33–48% evolution of the gene by forming transgenic lines. Studies have also shown that during glasshouse inoculation CRISPR system does not provide effective virus resistance.Citation112
CRISPR/Cas9-mediated gene insertion and replacement techniques have majorly developed herbicide-resistant crop varieties. In cassava, herbicide resistance was developed by HR and NHEJ DNA repair pathways by substituting crucial amino acids in the conserved domain of 5-enolpyruvylshikimate-3-phosphate synthase in order to give resistance against glyphosate-based herbicides (EPSPS)Citation117 and produce phenotypically normal glyphosate tolerant cassava. This method also provided knowledge about gene editing techniques for the further modification and improvement in cassava.Citation118 Recent studies have also shown that many attempts were made to develop cassava that can be used to visualize the early stages of CBB infection in vivo.Citation119 CRISPR-mediated homology-directed repair (HDR) was also used to generate plants with scarless insertion of GFP at the 3’ end of the CBB susceptibility (S) gene MeSWEET10a. These are successfully visualized at transcriptional and translational stages.
5.2. Trait Improvement via CRISPR/Cas9
In cassava, two isoforms of MESSIII genes, MESSIII-1 and MESSIII-2, were mutated simultaneously using CRISPR/Cas9 system, resulting in cassava with edited genes related to the starch synthesis pathway.Citation110 This research provides a platform to examine the role of genes in regulating amylopectin glucan synthesis in cassava. Studies have also shown that two amylose synthesis genes, PTST1 and GBSS, can reduce or remove amylose content in root starch by using CRISPR/Cas9-mediated targeted mutagenesis. Flowering in cassava was also accelerated by incorporating the Arabidopsis FLOWERING LOCUS T gene into the genome editing cassette, which was generally unusual in glasshouse conditions.Citation120 Researchers also achieved activated acceleration of cassava flowering in cassava by using CRISPR/Cas9 mediated disruption of Multiple TFL-like Floral Repressors.Citation121
During cotyledon-stage somatic embryogenesis, the mutants were phenotypically albino, which provided a good idea about gene’s role in plants using CRISPR/Cas9-mediated genome editing technology to target the phytoene desaturase (MePDS) gene in cassava. Using CRISPR/Cas9-mediated genome editing technology to target the phytoene desaturase (MePDS) gene in cassava, researchers could determine the relevance of the gene in the plant due to the albino phenotype of mutants during cotyledon-stage somatic embryogenesis. This prevented the requirement for gene sequencing to prove that a mutation had happened in the target gene.Citation122 Therefore, it provided a helpful arena for testing and enhancing CRISPR/Cas9 and other genome editing methods in cassava.
6. Different Strategies to Domesticate Orphan Crops to Develop Future-ready Crops
Most of the world’s population depends on a few species of crops like rice, maize, wheat, potato, and soy to meet the food and nutritional demands, so research and breeding efforts are mainly focused on these crop varieties. Some varieties of tasty, nutritious, and well-adapted orphan crops are also grown. Still, their cultivation is limited cause of their wild characteristics and can be domesticated by following strategic steps (). Many attempts were made to explore the domestication of these orphan crops, most suitable for genome editing techniques. Genes of P. pruinosa (groundcherry), an orphan crop, are modified to explore its domestication, especially the modification of those genes carried out whose orthologues control the domestication traits in the close relatives.Citation108 The results showed the power of this approach and demonstrated the importance of identifying mechanisms and gene targets. The path of domestication from wild ancestors to modern crops has become possible for several cultivated species with the help of genetic and genomic analysis.Citation102 Studies have shown that mutations that alter the functions of a few selected loci, known as domestication genes, have played a determining role. Wild teosinte and modern maize alleles at a few major loci are responsible for much of the difference.Citation102 Several domestication genes control flowering and fruit development, inhibiting the abscission of fruits and thus facilitating harvesting, increasing harvest index (more product per plant), and making the final product easier to store, chew, and digest have been found in different crop species. Manipulating these traits has proven an outstanding achievement in revolutionizing the domestication of different crop species.Citation102 Neolithic gatherers selected other traits like loss-of-shattering unintentionally. Increased food availability and domestication of crops enabled the flourishing of sciences, arts, and technology. Flourishing of sciences, arts and technology have emerged by the increase in food availability and domestication of crops. Most of the improvements of our stable crops are based on Neolithic selection, but recently other improvements are also seen, like semi-dwarfism in wheat and rice. The combination of spontaneous mutation in SELF-PRUNING in tomato was radically altered to enable its mechanical harvesting.
Figure 4. Showing the stepwise strategies to popularize the orphan crops for food and nutritional security.

The availability of genomic information and efficient genome editing tools played an important role in exploring crop domestication and improvement.Citation18,Citation102 Through these modifications, wild species and unimproved crops can also be modified in a targeted manner to produce novel and improved crops. An orphan crop groundcherry (P. pruinosa), which belongs to the solanaceous species, produces small but tasty berries. Due to its wild characteristics like sprawling habit, husked and small fruit, and strong fruit abscission, it cannot be grown on an agricultural scale.Citation123 Researchers saw an opportunity: is it possible to achieve the corresponding gains in this sister species by modifying the known gene targets of tomato domestication. They increased the number of flowers and delimited flowering time on both primary and axillary shoots by targeting repressors of the florigen pathway by gene editing techniques.Citation108 Knocking out many genes has shown great results, like knocking out a classical improvement gene; SELF-PRUNING, which controls indeterminate versus determinate growth in tomato, has proven very beneficial and resulted in extreme compactness. Knockout of another gene, SP5G, a florigen repressor, has increased axillary flowering and fruit destiny, although it does not affect a primary shoot.Citation108 Scientists also targeted the pathway which regulates shoot apical meristem size by the interaction of CLV3, a small peptide with its receptors (CLV1 and others) known as the CLAVATA pathway. Knocking out of CLV1 had shown effective results, viz. flower meristem size was increased, additional flower organs and two-locule fruit was converted to larger, three-locule fruit. These modifications generate variants most suited to full agricultural exploitations. These manipulations also gave a great idea about the possibilities of combining genomics and gene editing of just a small number of loci.Citation108 These studies also demonstrate the challenges that ‘domesticators will encounter.
Most important is to predict those targeted modifications that will generate the ideal phenotype. By understanding the domestication history of crops closely related to orphan crops, information about the target identity can be achieved. The structure of gene networks varies per node number, type, and connection.Citation124 Breeders and geneticists have identified that genetic modifiers present in the population can dramatically alter mutation phenotype. It was demonstrated by dwarfing effect in groundcherry by SELF PRUNING knockout or by the inability of SP5G manipulation to modify primary shoot flowering. Knockout may not be sufficient in some cases and may need subtler alleles like altering promoter activity or protein structure. Production of new alleles with new and valuable expression properties can be achieved by editing promoter segments.Citation106
Due to multigenic regulation of the same traits in orphan crops, their domestication is limited.Citation104 All things were considered that were important for taming of wild species. The work carried out by Lemmon et al. explained the approach’s feasibility and the importance of investing in research and enhanced information on genomes, genes, and cellular mechanisms behind plant traits. Some orphan crops serve as better candidates for gene manipulations than others, and some traits will be easier targets than others. This information and essential tools for manipulating DNA may serve as the ingredients for success. Accelerated domestication involves the manipulation of several genes with the connected combinatorial challenge of testing many variables.
Lastly, the faster domestication envisioned here may include the manipulation of several genes and the associated combinatorial difficulty of evaluating numerous factors. In reality, domesticated species may owe at least some of their success to their greater ease of genetic manipulation. If crucial domestication features were monogenic and variable in the progenitor, they would have been readily evident and selected by breeders. Optimistically, faster domestication will be an essential component of the survival toolkit, the collection of technologies required to sustain human civilization. At a minimum, understanding how domestication genes function in various animals will improve established crops.Citation104
7. Available Genetic Improvement Methods for New and Orphan Crops
7.1. Advanced and Conventional Breeding
The suitable approaches for genetic improvement and modification of any particularly new or orphan crop depend on the ideotype targets, good knowledge about inheritance, and genetic architecture of defining traits. The marker-assisted selection was widely used to develop major crops and has also begun to be applied to minor crops, that is, orphan crops. Examples of annual orphan crops include pigeon peaCitation26 and foxtail millet,Citation111 as also a wide range of perennial plants.Citation125,Citation126 However, in most cases, relatively high costs of phenotyping remain a constraint.Citation59
In most instances, the comparatively high costs of phenotyping continue to be a barrier.Citation59 This is especially true for perennial crops, whose evaluation requires several years of growth and whose huge life forms necessitate extensive area in field trials. Using genome-wide association scans, Cichy et al.Citation59 revealed genomic sites linked with variance in the so-called ‘cooking time characteristic’ in a common bean diversity panel.Citation127 Due to relatively substantial research expenditures, the common bean is arguably not precisely an orphan crop, but it is emblematic of other orphan legumes. The discovery of connections between some genomic regions and cooking time in legumes is significant since extended cooking durations restrict the seed’s efficiency as a food source.
Cichy et al.Citation59 found statistically significant associations between cooking time and single nucleotide polymorphisms (SNPs) on three Phaseolus vulgaris chromosomes, with the greatest connections seen on chromosome 6 (Pv06). Pv06 had two homologous cation/H+ exchanger genes, one homologous to AtCHX3 and the other to AtCHX4. On cassava (Manihot esculenta), a vegetatively propagated orphan annual root crop, the efficiency of genomic selection is now being studied.Citation128
Finger millet (Eleusine coracana), a seed-propagated annual grain, is the second example of an orphan crop for which this method is being investigated. As with conventional marker-assisted selection, the lack of phenotypic data from suitable training populations is the most significant barrier to applying genomic selection to novel and orphan crops.Citation59 However, implementing this strategy might be particularly beneficial for slow-maturing perennial novel and orphan crops that are difficult to phenotype directly for crucial production features.Citation125 When the underlying biological basis of crucial features is poorly understood, as with many novel and orphan crops, genomic selection may be very successful.
7.2. Speed Breeding
Another approach now being applied to orphan crops is speed breeding, which reduces the generation interval in breeding programs by modifying the photoperiod exposure of daylength-sensitive plants to accelerate their development (typically by prolonging ‘long-day’ plants’ exposure to light.Citation129 Each year, the number of probable generations has been extended from three to six for the long-day annual legume chickpea (Cicer arietinum).Citation130 Annual grain amaranth crops with “short days” have also proven successful.Citation131 Speed breeding should be especially beneficial when used with genomic selection, since this enables for selection during rapid cycling when complete phenotypic data is unavailable.Citation132 Transportable ‘speed breeding capsules,’ consisting of shipping containers retrofitted with temperature and light controls, irrigation systems, and greenhouse benches, have been proposed to lower the expenses of the speed breeding strategy for novel and orphan crops in low-income countries.Citation9
7.3. Participatory Breeding and Selection
In high-income countries, ‘citizen science’ programs that analyze crop germplasm have proven effective, as demonstrated by,Citation133 who investigated genotype-environment interactions in soybean (Glycine max) using data gathered by 1800 gardeners across Germany. Low-income countries, with larger rural populations and many engaged small-scale farmers, have an even greater opportunity for participatory experimentation. These communities may meaningfully analyze genetic materials in various target habitats and cropping systems and give additional information on crop yield and consumption if they are appropriately maintained. Participatory domestication approaches, for example, have been effectively used to genetically develop novel and orphan fruit tree crops in Central Africa, such as the semi-domesticated safou (Dacryodes edulis) and the incipiently domesticated bush mango (Irvingia gabonensis and I. wombolu).Citation133 The participative technique is particularly beneficial in low-income countries, where production circumstances are variable, and crop preferences are little recognized.Citation134 Both of these circumstances frequently apply to new and orphan crops.
Environmental considerations ‘Landscape genomic’ methods for crop growth are particularly important for perennial plants, which are largely wild populations that have evolved to local abiotic circumstances over many generations.Citation135 This is because the ‘in situ’ decision-making process saves time and effort compared to traditional field testing. Genomic data from plants growing in wild populations is connected with environmental factors using statistical approaches that account for the underlying adaptively neutral genetic structure induced by genetic drift.Citation136 Established correlations may be theoretically utilized to screen larger germplasm panels for favorable allele compositions for specific production circumstances. The enormous amount of georeferenced interpolated environmental data sets currently available digitally, such as temperature and precipitation profilesCitation137 and soil types, make comparisons easierCitation138,Citation139; for example, using soil maps to identify the soil environment as a critical driver of adaption in a landscape genomic analysis covering the native range of barrel medic (Medicago truncatula, a legume), with a high number of SNPs associated with soil variables, including SNPs in candidate genes involved in nodulation/symbiotic nitrogen fixation. The landscape method may be used for orphan crop landraces and new and orphan crops’ wild germplasm if local adaptation is presumed to have happened throughout orphan crop development and ecogeographical range expansion. A meta-analysis of various crop progenitors and landraces in the same geographical location might give comparative insights into natural and human adaptation mechanisms in this scenario. Statistical methods that link the results of many common garden genome-wide association studies, which look into the genetic basis of phenotype–trial site associations, with wild and/or landrace sample environment–genomic correlations are now available.Citation140 This will aid in the knowledge of causative loci for adaptability and the development of appropriate tactics for new and orphan crop range extension.
8. Engineering Insect-Pest Resistance through Exploiting Crop Wild Relatives
Studies on crop wild relatives (CWRs) have explained the loss of resilience traits that had occurred during the process of crop domestication (manmade selection) and is also known as “domestication syndrome,” according to which human needs were fulfilled by the developing crops which in turn made these crops susceptible and were not able to combat environmental fluctuations. According to these studies, most germplasm-cultivated lack the ability to manage insect attacks.Citation141 However, breeding for insect resistance using CWRs may prove a feasible strategy to increase the genetic diversity of the primary gene pool. Still, it has limitations as it remained unsuccessful in most of the crops due to the following reasons such as biological barriers, cross-incompatibility, linkage drag, and sterile embryo production. Different approaches like advanced genetic engineering tools such as transgenesis and genome editing were mainly used to introduce novel traits from CWRs into cultivated species. One of the essential aspects that are responsible for insect resistance is the identification of genetic variation. Comparative transcriptome and proteome analysis in response to insect feeding has proven the effective strategy for the identification of sequence and expression level polymorphism, where there is no feasibility of introgression. A tangible approach to introducing variability in the cultivated crops can be made by editing their genes which is based on the respective variation in CWRs, and it can be made feasible by using multi-omic strategies, firstly evaluating the variation in the sequences of relevant insect-responsive genes between the susceptible cultivated germplasm and the resistant wild relative. These can be successfully utilized for genome editing after validating resistance genes against the relevant pests. These approaches have successfully provided resistance in the cultivated gene pool to combat insect pests. Increasing research activities provided a platform for genome editing for insect pest management.Citation141,Citation142, Resistant phenotypes using sequence variation in economically important crops can be developed using overexpression or silencing strategies.Citation142 However, developing resistance via genome-editing-based sequence variation has not yet been proved.
Crop wild relatives include the progenitors of crops and other species that are more or less closely related to them. They provided plant breeders with a broad pool of potentially valuable genetic resources, thus proving beneficial to modern agriculture. Significant advances have been made after the 20 years of the Prescott-Allens’ study, which was based on molecular technologies and hybridization procedures available for breeding and cultivar development, that allowed the incorporation of more distantly-related taxa, and in our knowledge of the wild relatives available for use in these programs. The beneficial traits conferred by CWR genes included over 80% associated with pest and disease resistance. For over a century, breeders used wild relatives to get resistance to diseasesCitation143 and also searched extended gene pools for the genes that would provide resistance to major crop pests and diseases.Citation144,Citation145, They incorporated wild genes into about 13 crops, and all except barley and chickpea became disease resistant after the incorporation of wild genes. Since then, the use of this approach has increased steadily. Wild resistant traits were also used in tomatoes to develop disease resistance. It has been reported that this approach has been used at a rate of about one per year since 1982.Citation138 It has also been proven that the disease-resistant genes currently used in commercial cultivars have been bred from wild genetic resources (D. Zamir, personal communication). Fungicides have limited effectiveness against the pathogen, so growing lettuce in many parts of Europe may not be possible without introducing genes. However, resistance genes overcome these problems rapidly, and the breeders constantly return to wild germplasm for new resistance genes.Citation146,Citation147
9. Success Achieved in the Breeding of Orphan Crops
Since the beginning of the twenty-first century, unprecedented changes have occurred in plant breeding techniques. Some techniques that have contributed to the modernization of some of the crops in some regions include genotyping and phenotyping technologies, genomics, and analytics. Major crops have significantly contributed to the needed global increase in agricultural production, while orphan crops are global and of localized importance. The reality of orphan crops was that their research was limited. Also, there was very little knowledge available for orphan crops in genetics and genomics. Still, day by day, technologies are becoming more affordable and thus decreasing the cost of knowledge generation, often evolving the reality. Several international initiatives that were available for breeders, like the Generation Challenge Programme (GCP, www.generationcp.org/sunsetblog),Citation27 provided breeders access to genetic and genomic technologies for some orphan crops and also the knowledge previously available only for large commercial crops.Citation26 Different achievements seen in different crop species include characterization of genetic diversity,Citation148 understanding the basis of the genetics of agronomic traits,Citation149 and identification of elite alleles at target genesCitation150 for introgression in elite germplasm to impact crop performance.Citation151 Advancements in sequencing orphan-crop genomes have been significantly achieved over the last decadeCitation61 and also focus on developing tools aimed at discovering and characterizing loci and genes of use in molecular breeding of those sequenced species.Citation152
10. Digitalizing Breeding and Providing Support
Many attempts were made to modernize breeding techniques, and the modernizing approaches without a reliable data management system are very risky. Several research projects and breeding programs failed to achieve their goals due to poor data quality, lack of documentation, or lost institutional memory. Digitalization of breeding has brought several improvements, such as increased effectiveness of seed management, data capture, quality control, documentation, and analysis. More accuracy can be achieved in the selection decisions at all stages of the breeding process.Citation152 It also contributed to establishing routines to standardize the storage of germplasm information that include pedigree, phenotypes and genotypes, breeding protocols, metadata (location, climate, etc.), and trait oncology, which in turn provide different benefits such as enabling data mining and also sharing of opportunities across a broader range of environments and teams.Citation153
11. Conclusions and Future Outlook
Recalling orphan crops, negligible and underutilized crops that are not frequently operated globally but often play greater agricultural roles more in the area. These crops would deliver not only a more varied food organization less susceptible to to climate-induced inadequacies but also nourishing and stable food alternatives for the future. Genome editing of orphan crops could explore the domestication of these species. Domesticating an orphan crop plant requires multiple tools: a well-elucidated genome sequence, including the understanding of paralogue structure and gene expression, and a delivery system for genome editing, the simplest being a transformation system. Orphan crops have recently gained prominence for being stress tolerant and nutritious. Apart from being nutritious, these orphan crops can be grown with little agricultural input. Also, being stress-resilient, these crops are safe from the various vagaries of climate change and ensure yield even when other major crop varieties fail. Genome editing holds great promise for increasing crop productivity. There is particular interest in advancing breeding in orphan crops, which are often burdened by undesirable characteristics resembling wild relatives – developing genomic resources and efficient transformation methods in the orphan crops and using CRISPR/Cas9 to mutate orthologues of crop domestication and improve genes that control plant architecture, flower production, and fruit size, thereby improving these major productivity traits. Thus, translating knowledge from model crops enables the rapid creation of targeted allelic diversity and novel breeding germplasm in distantly related orphan crops. Genome editing could be used as a novel platform by plant breeders to speed up the domestication of semi-domesticated or even wild plants, creating a more diverse base for the sustainable provision of food and fodder in the future. Further domestication of underutilized crops may pave the way for rapid global food security and higher crop yields.
Author contributions
HY, RAM, AT, and BAB conceived the idea. HY, RAM, AT, BAB, KAB, IBN, and AR, collected the literature and participated in writing. ID, PVVP, RAM and AR proofread and edited the final version. All authors have read and approved the final version of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Dawson IK, Jaenicke H. Underutilised plant species: the role of biotechnology. Position Paper No. 1. Crops for the Future; 2006.
- Umesh MR, Angadi S, Gowda P, Ghimire R, Begna S. Climate-resilient minor crops for food security. In Agronomic crops. Singapore: Springer; 2019. pp. 19–32.
- Hendre PS, Muthemba S, Kariba R, Muchugi A, Fu Y, Chang Y, Song B, Liu H, Liu M, Liao X, et al. African Orphan Crops Consortium (AOCC): status of developing genomic resources for African orphan crops. Planta. 2019;250(3):989–1003. doi:10.1007/s00425-019-03156-9.
- Tadele Z, Bartels D. Promoting orphan crops research and development. Germany: Springer-Verlag GmbH; 2019. p. 675–76.
- Gregory PJ, Mayes S, Hui CH, Jahanshiri E, Julkifle A, Kuppusamy G, Kuan HW, Lin TX, Massawe F, Suhairi TASTM, et al. Crops for the future (CFF): an overview of research efforts in the adoption of underutilised species. Planta. 2019;250(3):979–88. doi:10.1007/s00425-019-03179-2.
- Dawson IK, Powell W, Hendre P, Bančič J, Hickey JM, Kindt R, Hoad S, Hale I, Jamnadass R. The role of genetics in mainstreaming the production of new and orphan crops to diversify food systems and support human nutrition. New Phytologist. 2019;224(1):37–54. doi:10.1111/nph.15895.
- Oibiokpa FI, Adoga GI, Saidu AN, Kudirat OS. Nutritional composition of detarium microcarpum fruit. African Journal of Food Science. 2014;8(6):342–50. doi:10.5897/AJFS2014.1161.
- Cullis C, Kunert KJ. Unlocking the potential of orphan legumes. J Exp Bot. 2017;68(8):1895–903. doi:10.1093/jxb/erw437.
- Chiurugwi T, Kemp S, Powell W, Hickey LT. Speed breeding orphan crops. Theor Appl Genet. 2019;132(3):607–16. doi:10.1007/s00122-018-3202-7.
- Mabhaudhi T, Chimonyo VGP, Hlahla S, Massawe F, Mayes S, Nhamo L, Modi AT. Prospects of orphan crops in climate change. Planta. 2019;250(3):695–708. doi:10.1007/s00425-019-03129-y.
- Borelli T, Hunter D, Padulosi S, Amaya N, Meldrum G, de Oliveira Beltrame DM, Samarasinghe G, Wasike VW, Güner B, Tan A, et al. Local solutions for sustainable food systems: the contribution of orphan crops and wild edible species. Agronomy. 2020;10(2):231. doi:10.3390/agronomy10020231.
- King AJ, Montes LR, Clarke JG, Itzep J, Perez CAA, Jongschaap REE, Visser RGF, van Loo EN, Graham IA. Identification of QTL markers contributing to plant growth, oil yield and fatty acid composition in the oilseed crop Jatropha curcas L. Biotechnol Biofuels. 2015;8(1):1–17. doi:10.1186/s13068-015-0326-8.
- Tlili N, Elfalleh W, Saadaoui E, Khaldi A, Triki S, Nasri N. The caper (Capparis L.): ethnopharmacology, phytochemical and pharmacological properties. Fitoterapia. 2011;82(2):93–101. doi:10.1016/j.fitote.2010.09.006.
- Saikia JP, Konwar BK. Physicochemical properties of starch from aroids of North East India. Int J Food Prop. 2012;15(6):1247–61. doi:10.1080/10942912.2010.491929.
- Tolera A, Sundstøl F. Supplementation of graded levels of Desmodium intortum hay to sheep feeding on maize stover harvested at three stages of maturity: 1. Feed intake, digestibility and body weight change. Anim Feed Sci Technol. 2000;85(3–4):239–57. doi:10.1016/S0377-8401(00)00135-8.
- Yu H, Lin T, Meng X, Du H, Zhang J, Liu G, Chen M, Jing Y, Kou L, Li X, et al. A route to de novo domestication of wild allotetraploid rice. Cell. 2021;184(5):1156–70. doi:10.1016/j.cell.2021.01.013.
- Karavolias NG, Horner W, Abugu MN, Evanega SN. Application of gene editing for climate change in agriculture. Front Sustain Food Syst. 2021;5:685801. doi:10.3389/fsufs.2021.685801.
- Østerberg JT, Xiang W, Olsen LI, Edenbrandt AK, Vedel SE, Christiansen A, Landes X, Andersen MM, Pagh P, Sandøe P, et al. Accelerating the domestication of new crops: feasibility and approaches. Trends Plant Sci. 2017;22(5):373–84. doi:10.1016/j.tplants.2017.01.004.
- Padulosi S, King EI, Hunter D, Swaminathan MS, editors. Orphan Crops for Sustainable Food and Nutrition Security: Promoting Neglected and Underutilized Species. London (UK): Routledge; 2021.
- Talabi AO, Vikram P, Thushar S, Rahman H, Ahmadzai H, Nhamo N, Shahid M, Singh RK. Orphan crops: a best fit for dietary enrichment and diversification in highly deteriorated marginal environments. Front Plant Sci. 2022;13. doi:10.3389/fpls.2022.839704.
- Raskin I, Ribnicky DM, Komarnytsky S, Ilic N, Poulev A, Borisjuk N, Brinker A, Moreno DA, Ripoll C, Yakoby N, et al. Plants and human health in the twenty-first century. Trends Biotechnol. 2002;20(12):522–31. doi:10.1016/S0167-7799(02)02080-2.
- Hüdig M, Laibach N, Hein A-C. Genome editing in crop plant research—alignment of expectations and current developments. Plants. 2022;11(2):212. doi:10.3390/plants11020212.
- Li T, Yang X, Yu Y, Si X, Zhai X, Zhang H, Dong W, Gao C, Xu C. Domestication of wild tomato is accelerated by genome editing. Nat Biotechnol. 2018;36(12):1160–63. doi:10.1038/nbt.4273.
- Zsögön A, Čermák T, Naves ER, Notini MM, Edel KH, Weinl S, Freschi L, Voytas DF, Kudla J, Peres LEP, et al. De novo domestication of wild tomato using genome editing. Nat Biotechnol. 2018;36(12):1211–16. doi:10.1038/nbt.4272.
- Shewry PR, Tatham AS. Improving wheat to remove coeliac epitopes but retain functionality. J Cereal Sci. 2016;67:12–21. doi:10.1016/j.jcs.2015.06.005.
- Varshney RK, Chen W, Li Y, Bharti AK, Saxena RK, Schlueter JA, Donoghue MTA, Azam S, Fan G, Whaley AM, et al. Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource-poor farmers. Nat Biotechnol. 2012;30(1):83. doi:10.1038/nbt.2022.
- Ribaut J-M, Ragot M. Modernising breeding for orphan crops: tools, methodologies, and beyond. Planta. 2019;250(3):971–77. doi:10.1007/s00425-019-03200-8.
- Cannarozzi G, Plaza-Wüthrich S, Esfeld K, Larti S, Wilson YS, Girma D, de Castro E, Chanyalew S, Blösch R, Farinelli L, et al. Genome and transcriptome sequencing identifies breeding targets in the orphan crop tef (Eragrostis tef). BMC Genomics. 2014;15(1):1–21. doi:10.1186/1471-2164-15-581.
- Doust A, Diao X. Genetics and genomics of Setaria. Switzerland (AG): Springer Nature; 2017. Vol. 19.
- Hittalmani S, Mahesh HB, Shirke MD, Biradar H, Uday G, Aruna YR, Lohithaswa HC, Mohanrao A. Genome and transcriptome sequence of finger millet (Eleusine coracana (L.) Gaertn.) provides insights into drought tolerance and nutraceutical properties. BMC Genomics. 2017;18(1):1–16. doi:10.1186/s12864-017-3850-z.
- C-Y Y, Fan L. Orphan crops and their wild relatives in the genomic era. Mol Plant. 2021;14(1):27–39. doi:10.1016/j.molp.2020.12.013.
- Linares OF. African rice (Oryza glaberrima): history and future potential. Proc Natl Acad Sci. 2002;99(25):16360–65. doi:10.1073/pnas.252604599.
- Caballero B, Finglas PM, Toldra F. Encyclopedia of food and health. In: Jag AA, Accc C-M, RAM S, editors. Brazil: Universidade de Sao Paulo. Sao Paulo. Elsevier Ltd; 2016.
- Mayes S, Ho WK, Chai HH, Gao X, Kundy AC, Mateva KI, Zahrulakmal M, Hahiree MKIM, Kendabie P, Licea LCS, et al. Bambara groundnut: an exemplar underutilised legume for resilience under climate change. Planta. 2019;250(3):803–20. doi:10.1007/s00425-019-03191-6.
- Singh M, Upadhyaya HD. Genetic and genomic resources for grain cereals improvement. USA: Elsevier; 2015.
- Wijngaard H, Arendt EK. Buckwheat. Cereal Chemistry. 2006;83(4):391–401. doi:10.1094/CC-83-0391.
- Ceballos H, Iglesias CA, Pérez JC, Dixon AGO. Cassava breeding: opportunities and challenges. Plant Mol Biol. 2004;56(4):503–16. doi:10.1007/s11103-004-5010-5.
- Mafakheri A, Siosemardeh AF, Bahramnejad B, Struik PC, Sohrabi Y. Effect of drought stress on yield, proline and chlorophyll contents in three chickpea cultivars. Australian journal of crop science. 2010;4(8):580–5.
- Timko MP, Singh BB. Cowpea, a multifunctional legume. Genomics of tropical crop plants. New York (NY): Springer; 2008. p. 227–58.
- Olango TM, Tesfaye B, Catellani M, Pè ME. Indigenous knowledge, use and on-farm management of enset (Ensete ventricosum (Welw.) Cheesman) diversity in Wolaita, Southern Ethiopia. J Ethnobiol Ethnomed. 2014;10(1):1–18. doi:10.1186/1746-4269-10-41.
- Caballero B, Finglas P, Toldrá F. Encyclopedia of food and health. Waltham (MA): Academic Press; 2015.
- Girma D, Korbu L. Genetic improvement of grass pea (Lathyrus sativus) in Ethiopia: an unfulfilled promise. Plant Breeding. 2012;131(2):231–36. doi:10.1111/j.1439-0523.2011.01935.x.
- Singh M, Upadhyaya HD, Bisht IS. Genetic and genomic resources of grain legume improvement. Waltham (MA): Newnes; 2013.
- de la Vega MP, Torres AM, Cubero JI, Kole C, editors. Genetics, genomics and breeding of cool season grain legumes. UK: CRC Press; 2011.
- Zuk M, Richter D, Matuła J, Szopa J. Linseed, the multipurpose plant. Ind Crops Prod. 2015;75:165–77. doi:10.1016/j.indcrop.2015.05.005.
- Kumar A, Kumar P, Nadendla R. A review on: abelmoschus esculentus (Okra). Int J Pharm Sci Res. 2013;3:129–32.
- Yadav HP, Gupta SK, Rajpurohit BS, Pareek, N. Pearl millet. In: Broadening the genetic base of grain cereals. New Delhi: Springer; 2016. p. 205–24.
- Manickavasagan A, Thirunathan P. Pulses: processing and product development. Switzerland: Springer Nature; 2020.
- Kumar B, Bhalothia P. Orphan crops for future food security. J Biosci. 2020;45(1):1–8. doi:10.1007/s12038-020-00107-5.
- Atherton J, Rees A. Tropical root and tuber crops: cassava, sweet potato, yams and aroids. 2008;2:1–9. doi:10.1079/9781789243369.0000.
- Spaenij-Dekking L, Kooy-Winkelaar Y, Koning F. The Ethiopian cereal tef in celiac disease. N Engl J Med. 2005;353(16):1748–49. doi:10.1056/NEJMc051492.
- Raymundo R, Asseng S, Cammarano D, Quiroz R. Potato, sweet potato, and yam models for climate change: a review. Field Crops Res. 2014;166:173–85. doi:10.1016/j.fcr.2014.06.017.
- Farooq MS, Uzaiir M, Raza A, Habib M, Xu Y, Yousuf M, Yang SH, Ramzan Khan M. Uncovering the research gaps to alleviate the negative impacts of climate change on food security: a review. Front Plant Sci. 2022;13:927535. doi:10.3389/fpls.2022.927535.
- Garibaldi LA, Gemmill-Herren B, D’Annolfo R, Graeub BE, Cunningham SA, Breeze TD. Farming approaches for greater biodiversity, livelihoods, and food security. Trends Ecol Evol. 2017;32(1):68–80. doi:10.1016/j.tree.2016.10.001.
- Rasmussen C, Lagnaoui A, Esbjerg P. Advances in the knowledge of quinoa pests. Food Rev Int. 2003;19(1–2):61–75. doi:10.1081/FRI-120018868.
- Habiyaremye C, Matanguihan JB, D’Alpoim Guedes J, Ganjyal GM, Whiteman MR, Kidwell KK, Murphy KM. Proso millet (Panicum miliaceum L.) and its potential for cultivation in the Pacific Northwest, US: a review. Front Plant Sci. 2017;7:1961. doi:10.3389/fpls.2016.01961.
- Bohra A, Tiwari A, Kaur P, Ganie SA, Raza A, Roorkiwal M, Mir RR, Fernie AR, Smýkal P, Varshney RK, et al. The key to the future lies in the past: insights from grain legume domestication and improvement should inform future breeding strategies. Plant Cell Physiol. 2022. doi:10.1093/pcp/pcac086.
- Raza A, Tabassum J, Fakhar AZ, Sharif R, Chen H, Zhang C, Ju L, Fotopoulos V, Siddique KHM, Singh RK, et al. Smart reprograming of plants against salinity stress using modern biotechnological tools. Crit Rev Biotechnol. 2022;1–28 . doi:10.1080/07388551.2022.2093695
- Varshney RK, Ribaut J-M, Buckler ES, Tuberosa R, Rafalski JA, Langridge P. Can genomics boost productivity of orphan crops? Nat Biotechnol. 2012;30(12):1172–76. doi:10.1038/nbt.2440.
- Varshney RK, Bohra A, Yu J, Graner A, Zhang Q, Sorrells ME. Designing future crops: genomics-assisted breeding comes of age. Trends Plant Sci. 2021;26(6):631–49. doi:10.1016/j.tplants.2021.03.010.
- Varshney RK, Close TJ, Singh NK, Hoisington DA, Cook DR. Orphan legume crops enter the genomics era! Current opinion in plant biology. Current Opinion in Plant Biology. 2009;12(2):202–10. doi:10.1016/j.pbi.2008.12.004.
- Tadele Z. Orphan crops: their importance and the urgency of improvement. Planta. 2019;250(3):677–94. doi:10.1007/s00425-019-03210-6.
- Jackson SA, Iwata A, Lee SH, Schmutz J, Shoemaker R. Sequencing crop genomes: approaches and applications. New Phytologist. 2011;191(4):915–25. doi:10.1111/j.1469-8137.2011.03804.x.
- Olsen KM, Wendel JF. A bountiful harvest: genomic insights into crop domestication phenotypes. Annu Rev Plant Biol. 2013;64(1):47–70. doi:10.1146/annurev-arplant-050312-120048.
- Díaz A, Zikhali M, Turner AS, Isaac P, Laurie DA. Copy number variation affecting the Photoperiod-B1 and Vernalization-A1 genes is associated with altered flowering time in wheat (Triticum aestivum). PloS one. 2012;7(3):e33234. doi:10.1371/journal.pone.0033234.
- Knox AK, Dhillon T, Cheng H, Tondelli A, Pecchioni N, Stockinger EJ. CBF gene copy number variation at frost resistance-2 is associated with levels of freezing tolerance in temperate-climate cereals. Theor Appl Genet. 2010;121(1):21–35. doi:10.1007/s00122-010-1288-7.
- Morrell PL, Buckler ES, Ross-Ibarra J. Crop genomics: advances and applications. Nat Rev Genet. 2012;13(2):85–96. doi:10.1038/nrg3097.
- Salman-Minkov A, Sabath N, Mayrose I. Whole-genome duplication as a key factor in crop domestication. Nature Plants. 2016;2(8):1–4. doi:10.1038/nplants.2016.115.
- Wu J, Wang Z, Shi Z, Zhang S, Ming R, Zhu S, Khan MA, Tao S, Korban SS, Wang H, et al. The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res. 2013;23(2):396–408. doi:10.1101/gr.144311.112.
- Clouse JW, Adhikary D, Page JT, Ramaraj T, Deyholos MK, Udall JA, Fairbanks DJ, Jellen EN, Maughan PJ. The amaranth genome: genome, transcriptome, and physical map assembly. Plant Genome. 2016;9(1):lantgenome2015–07. doi:10.3835/plantgenome2015.07.0062.
- Regalado JJ, Tossi VE, Burrieza HP, Encina CL, Pitta-Alvarez SI. Micropropagation protocol for coastal quinoa. Plant Cell, Tissue and Organ Culture (PCTOC). 2020;142(1):213–19. doi:10.1007/s11240-020-01840-3.
- Wang W, Feng B, Xiao J, Xia Z, Zhou X, Li P, Zhang W, Wang Y, Møller BL, Zhang P, et al. Cassava genome from a wild ancestor to cultivated varieties. Nat Commun. 2014;5(1):1–9.
- Yang MM, Wang J, Dong L, Kamizaki K, Wang Z, Tamada K, Takumi T, Hashimoto R, Otani H, Pazour GJ, et al. Lack of association of C3 gene with uveitis: additional insights into the genetic profile of uveitis regarding complement pathway genes. Sci Rep. 2017;7(1):1–8. doi:10.1038/s41598-016-0028-x.
- Siadjeu C, Pucker B, Viehöver P, Albach DC, Weisshaar B. High contiguity de novo genome sequence assembly of trifoliate yam (Dioscorea dumetorum) using long read sequencing. Genes. 2020;11(3):274. doi:10.3390/genes11030274.
- Saeed F, Chaudhry UK, Bakhsh A, Raza A, Saeed Y, Bohra A, Varshney RK. Moving beyond DNA sequence to improve plant stress responses. Front Genet. 2022;13:874648. doi:10.3389/fgene.2022.874648.
- Raza A. Metabolomics: a systems biology approach for enhancing heat stress tolerance in plants. Plant Cell Rep. 2022;41(3):741–63. doi:10.1007/s00299-020-02635-8.
- Raza A, Tabassum J, Zahid Z, Charagh S, Bashir S, Barmukh R, Khan RSA, Barbosa F, Zhang C, Chen H, et al. Advances in ‘omics’ approaches for improving toxic metals/metalloids tolerance in plants. Front Plant Sci. 2022;12:794373. doi:10.3389/fpls.2021.794373.
- Raza A, Razzaq A, Mehmood SS, Hussain MA, Wei S, He H, Zaman QU, Xuekun Z, Yong C, Hasanuzzaman M, et al. Omics: the way forward to enhance abiotic stress tolerance in Brassica napus L. GM Crops Food. 2021;12(1):251–81. doi:10.1080/21645698.2020.1859898.
- Raza A, Tabassum J, Kudapa H, Varshney RK. Can omics deliver temperature resilient ready-to-grow crops? Crit Rev Biotechnol. 2021;41(8):1209–32. doi:10.1080/07388551.2021.1898332.
- Raza A, Mubarik, MS, Sharif, R, Habib, M, Jabeen, W, Zhang, C, Chen, H, Chen, ZH, Siddique, KHM, Zhuang, W, Varshney, RK , Developing drought‐smart, ready‐to‐grow future crops. Plant Genome 2022;():e20279. doi:10.1002/tpg2.20279.
- L-m C, X-w L, T-j H, Li P-J, Liu Y, Zhou S-X, Wu Q-C, Chen -T-T, Lu Y-B, Hou Y-M, et al. Comparative biochemical and transcriptome analyses in tomato and eggplant reveal their differential responses to Tuta absoluta infestation. Genomics. 2021;113(4):2108–21. doi:10.1016/j.ygeno.2021.05.002.
- Bhardwaj AR, Joshi G, Kukreja B, Malik V, Arora P, Pandey R, Shukla RN, Bankar KG, Katiyar-Agarwal S, Goel S, et al. Global insights into high temperature and drought stress regulated genes by RNA-Seq in economically important oilseed crop Brassica juncea. BMC Plant Biol. 2015;15(1):1–15. doi:10.1186/s12870-014-0405-1.
- Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet. 2011;12(2):87–98. doi:10.1038/nrg2934.
- Ranasinghe R, Maduwanthi SDT, Marapana R. Nutritional and health benefits of jackfruit (Artocarpus heterophyllus Lam.): a review. Int J Food Sci. 2019;2019.
- Golisz A, Sugano M, Fujii Y. Microarray expression profiling of Arabidopsis thaliana L. in response to allelochemicals identified in buckwheat. J Exp Bot. 2008;59(11):3099–109. doi:10.1093/jxb/ern168.
- Zhu YY, Zeng HQ, Dong CX, Yin XM, Shen QR, Yang ZM. microRNA expression profiles associated with phosphorus deficiency in white lupin (Lupinus albus L.). Plant Sci. 2010;178(1):23–29. doi:10.1016/j.plantsci.2009.09.011.
- Schmidt S, Blom JF, Pernthaler J, Berg G, Baldwin A, Mahenthiralingam E, Eberl L. Production of the antifungal compound pyrrolnitrin is quorum sensing-regulated in members of the Burkholderia cepacia complex. Environ Microbiol. 2009;11(6):1422–37. doi:10.1111/j.1462-2920.2009.01870.x.
- Srivastava S, Srivastava AK, Sablok G, Deshpande TU, Suprasanna P. Transcriptomics profiling of Indian mustard (Brassica juncea) under arsenate stress identifies key candidate genes and regulatory pathways. Front Plant Sci. 2015;6:646. doi:10.3389/fpls.2015.00646.
- Manning K, Tör M, Poole M, Hong Y, Thompson AJ, King GJ, Giovannoni JJ, Seymour GB. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet. 2006;38(8):948–52. doi:10.1038/ng1841.
- Butelli E, Licciardello C, Zhang Y, Liu J, Mackay S, Bailey P, Reforgiato-Recupero G, Martin C. Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell. 2012;24(3):1242–55. doi:10.1105/tpc.111.095232.
- Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S-I, Natsume T, Takehana K, Yamada N, et al. Nutrient-dependent mTORC1 association with the ULK1–Atg13–FIP200 complex required for autophagy. Mol Biol Cell. 2009;20(7):1981–91. doi:10.1091/mbc.e08-12-1248.
- Chang Y, Liu H, Liu M, Liao X, Sahu SK, Fu Y, Song B, Cheng S, Kariba R, Muthemba S, et al. The draft genomes of five agriculturally important African orphan crops. GigaScience. 2019;8(3):giy152. doi:10.1093/gigascience/giy152.
- Ong-Abdullah M, Ordway JM, Jiang N, Ooi S-E, Kok S-Y, Sarpan N, Azimi N, Hashim AT, Ishak Z, Rosli SK, et al. Loss of Karma transposon methylation underlies the mantled somaclonal variant of oil palm. Nature. 2015;525(7570):533–37. doi:10.1038/nature15365.
- Varshney RK, Saxena RK, Upadhyaya HD, Khan AW, Yu Y, Kim C, Rathore A, Kim D, Kim J, An S, et al. Whole-genome resequencing of 292 pigeonpea accessions identifies genomic regions associated with domestication and agronomic traits. Nat Genet. 2017;49(7):1082–88. doi:10.1038/ng.3872.
- Tohge T, de Souza Lp, Fernie AR, de Souza LP. Current understanding of the pathways of flavonoid biosynthesis in model and crop plants. J Exp Bot. 2017;68(15):4013–28. doi:10.1093/jxb/erx177.
- Paszke A, Gross S, Massa F, Lerer A, Bradbury J, Chanan G, Killeen T, Lin Z, Gimelshein N, Antiga L, et al. Pytorch: an imperative style, high-performance deep learning library. Adv Neural Inf Process Syst. 2019;32.
- Klee HJ, Tieman DM. The genetics of fruit flavour preferences. Nat Rev Genet. 2018;19(6):347–56. doi:10.1038/s41576-018-0002-5.
- Beleggia R, Rau D, Laidò G, Platani C, Nigro F, Fragasso M, De Vita P, Scossa F, Fernie AR, Nikoloski Z, et al. Evolutionary metabolomics reveals domestication-associated changes in tetraploid wheat kernels. Mol Biol Evol. 2016;33(7):1740–53. doi:10.1093/molbev/msw050.
- Gapare W, Liu S, Conaty W, Zhu Q-H, Gillespie V, Llewellyn D, Stiller W, Wilson I. Historical datasets support genomic selection models for the prediction of cotton fiber quality phenotypes across multiple environments. G3: Genes, Genomes, Genetics. 2018;8(5):1721–32. doi:10.1534/g3.118.200140.
- Langner T, Kamoun S, Belhaj K. CRISPR crops: plant genome editing toward disease resistance. Annu Rev Phytopathol. 2018;56(1):479–512. doi:10.1146/annurev-phyto-080417-050158.
- Yin K, Gao C, Qiu J-L. Progress and prospects in plant genome editing. Nature Plants. 2017;3(8):1–6. doi:10.1038/nplants.2017.107.
- Doebley JF, Gaut BS, Smith BD. The molecular genetics of crop domestication. Cell. 2006;127(7):1309–21. doi:10.1016/j.cell.2006.12.006.
- Čermák T, Curtin SJ, Gil-Humanes J, Čegan R, Kono TJY, Konečná E, Belanto JJ, Starker CG, Mathre JW, Greenstein RL, et al. A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell. 2017;29(6):1196–217. doi:10.1105/tpc.16.00922.
- Diamond J. Evolution, consequences and future of plant and animal domestication. Nature. 2002;418(6898):700–07. doi:10.1038/nature01019.
- Valle‐Echevarria AD, Fumia N, Gore MA, Kantar M. Accelerating crop domestication in the era of gene editing. Plant Breed Rev. 2021;45:185–211.
- Rodríguez-Leal D, Lemmon ZH, Man J, Bartlett ME, Lippman ZB. Engineering quantitative trait variation for crop improvement by genome editing. Cell. 2017;171(2):470–80. doi:10.1016/j.cell.2017.08.030.
- Falke KC, Glander S, He F, Hu J, de Meaux J, Schmitz G. The spectrum of mutations controlling complex traits and the genetics of fitness in plants. Curr Opin Genet Dev. 2013;23(6):665–71. doi:10.1016/j.gde.2013.10.006.
- Lemmon ZH, Reem NT, Dalrymple J, Soyk S, Swartwood KE, Rodriguez-Leal D, Van Eck J, Lippman ZB. Rapid improvement of domestication traits in an orphan crop by genome editing. Nature Plants. 2018;4(10):766–70. doi:10.1038/s41477-018-0259-x.
- Zsögön A, Peres LEP, Xiao Y, Yan J, Fernie AR. Enhancing crop diversity for food security in the face of climate uncertainty. Plant J. 2022;109(2):402–14. doi:10.1111/tpj.15626.
- Zhao M, Tang S, Zhang H, He M, Liu J, Zhi H, Sui Y, Liu X, Jia G, Zhao Z, et al. DROOPY LEAF1 controls leaf architecture by orchestrating early brassinosteroid signaling. Proc Natl Acad Sci. 2020;117(35):21766–74. doi:10.1073/pnas.2002278117.
- Mamidi S, Healey A, Huang P, Grimwood J, Jenkins J, Barry K, Sreedasyam A, Shu S, Lovell JT, Feldman M, et al. A genome resource for green millet Setaria viridis enables discovery of agronomically valuable loci. Nat Biotechnol. 2020;38(10):1203–10. doi:10.1038/s41587-020-0681-2.
- Bull SE, Seung D, Chanez C, Mehta D, Kuon J-E, Truernit E, Hochmuth A, Zurkirchen I, Zeeman SC, Gruissem W, et al. Accelerated ex situ breeding of GBSS - and PTST1 -edited cassava for modified starch. Sci Adv. 2018;4(9):eaat6086. doi:10.1126/sciadv.aat6086.
- Prochnik S, Marri PR, Desany B, Rabinowicz PD, Kodira C, Mohiuddin M, Rodriguez F, Fauquet C, Tohme J, Harkins T, et al. The cassava genome: current progress, future directions. Trop Plant Biol. 2012;5(1):88–94. doi:10.1007/s12042-011-9088-z.
- Cassava NNMA. Manihot esculenta Crantz, genetic resources: origin of the crop, its evolution and relationships with wild relatives. Genet Mol Res. 2002;1:298–305.
- Malik AI, Kongsil P, Nguyễn VA, Ou W, Srean P, López-Lavalle LA, Utsumi Y, Lu C, Kittipadakul P, Nguyễn HH, et al. Cassava breeding and agronomy in Asia: 50 years of history and future directions. Breed Sci. 2020;70:18180.
- Gomez MA, Lin ZD, Moll T, Chauhan RD, Hayden L, Renninger K, Beyene G, Taylor NJ, Carrington JC, Staskawicz BJ, et al. Simultaneous CRISPR/Cas9-mediated editing of cassava eIF4E isoforms nCBP-1 and nCBP-2 reduces cassava brown streak disease symptom severity and incidence. Plant Biotechnol J. 2019;17(2):421–34. doi:10.1111/pbi.12987.
- Hummel AW, Chauhan RD, Cermak T, Mutka AM, Vijayaraghavan A, Boyher A, Starker CG, Bart R, Voytas DF, Taylor NJ, et al. Allele exchange at the EPSPS locus confers glyphosate tolerance in cassava. Plant Biotechnol J. 2018;16(7):1275–82. doi:10.1111/pbi.12868.
- Hu Y, Li W, Gao T, Cui Y, Jin Y, Li P, Ma Q, Liu X, Cao C. The severe acute respiratory syndrome coronavirus nucleocapsid inhibits type I interferon production by interfering with TRIM25-mediated RIG-I ubiquitination. J Virol. 2017;91(8):e02143–16. doi:10.1128/JVI.02143-16.
- Veley KM, Okwuonu I, Jensen G, Yoder M, Taylor NJ, Meyers BC, Bart RS. Gene tagging via CRISPR-mediated homology-directed repair in cassava. G3. 2021;11(4):jkab028. doi:10.1093/g3journal/jkab028.
- Tyagi S, Kumar R, Kumar V, Won SY, Shukla P. Engineering disease resistant plants through CRISPR-Cas9 technology. GM Crops Food. 2021;12(1):125–44. doi:10.1080/21645698.2020.1831729.
- Odipio J, Alicai T, Nusinow D, Bart R, Taylor N, editors. CRISPR/Cas9-mediated disruption of multiple TFL1-like floral repressors activates flowering in cassava 2018: SPRINGER 233 SPRING ST, New York, NY 10013 USA.
- Odipio J, Alicai T, Ingelbrecht I, Nusinow DA, Bart R, Taylor NJ. Efficient CRISPR/Cas9 genome editing of phytoene desaturase in cassava. Front Plant Sci. 2017;8:1780. doi:10.3389/fpls.2017.01780.
- Comai L. The taming of the shrub. Nat Plants. 2018;4(10):742–43. doi:10.1038/s41477-018-0275-x.
- Alonso-Blanco C, Aarts MGM, Bentsink L, Keurentjes JJB, Reymond M, Vreugdenhil D, Koornneef M. What has natural variation taught us about plant development, physiology, and adaptation? Plant Cell. 2009;21(7):1877–96. doi:10.1105/tpc.109.068114.
- Isik F, Kumar S, Martínez-García PJ, Iwata H, Yamamoto T. Acceleration of forest and fruit tree domestication by genomic selection. In: Christophe P, Anne-Françoise Adam-Blondon, Editors. Advances in botanical research. USA: Elsevier; 2015, Vol. 74, p. 93–124.
- Migicovsky Z, Myles S. Exploiting wild relatives for genomics-assisted breeding of perennial crops. Front Plant Sci. 2017;8:460. doi:10.3389/fpls.2017.00460.
- Cichy KA, Wiesinger JA, Mendoza FA. Genetic diversity and genome-wide association analysis of cooking time in dry bean (Phaseolus vulgaris L.). Theor Appl Genet. 2015;128(8):1555–67. doi:10.1007/s00122-015-2531-z.
- Ozimati A, Kawuki R, Esuma W, Kayondo IS, Wolfe M, Lozano R, Rabbi I, Kulakow P, Jannink J-L. Training population optimization for prediction of cassava brown streak disease resistance in West African Clones. G3: Genes, Genomes, Genetics. 2018;8(12):3903–13. doi:10.1534/g3.118.200710.
- Ghosh S, Watson A, Gonzalez-Navarro OE, Ramirez-Gonzalez RH, Yanes L, Mendoza-Suárez M, Simmonds J, Wells R, Rayner T, Green P, et al. Speed breeding in growth chambers and glasshouses for crop breeding and model plant research. Nat Protoc. 2018;13(12):2944–63. doi:10.1038/s41596-018-0072-z.
- Watson A, Ghosh S, Williams MJ, Cuddy WS, Simmonds J, Rey M-D, Asyraf Md Hatta M, Hinchliffe A, Steed A, Reynolds D, et al. Speed breeding is a powerful tool to accelerate crop research and breeding. Nature Plants. 2018;4(1):23–29. doi:10.1038/s41477-017-0083-8.
- Stetter MG, Zeitler L, Steinhaus A, Kroener K, Biljecki M, Schmid KJ. Crossing methods and cultivation conditions for rapid production of segregating populations in three grain amaranth species. Front Plant Sci. 2016;7:816. doi:10.3389/fpls.2016.00816.
- Li H, Birol I. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34(18):3094–100. doi:10.1093/bioinformatics/bty191.
- Würschum T, Leiser WL, Jähne F, Bachteler K, Miersch M, Hahn V. The soybean experiment ‘1000 Gardens’: a case study of citizen science for research, education, and beyond. Theor Appl Genet. 2019;132(3):617–26. doi:10.1007/s00122-018-3134-2.
- Bhargav DK, Meena HP, Ppb PPB. Participatory plant breeding: farmers as breeders. Popular Kheti. 2014;2:7–14.
- Bragg JG, Supple MA, Andrew RL, Borevitz JO. Genomic variation across landscapes: insights and applications. New Phytologist. 2015;207(4):953–67. doi:10.1111/nph.13410.
- Coop G, Witonsky D, Di Rienzo A, Pritchard JK. Using environmental correlations to identify loci underlying local adaptation. Genetics. 2010;185(4):1411–23. doi:10.1534/genetics.110.114819.
- Fick SE, Hijmans RJ. WorldClim 2: new 1‐km spatial resolution climate surfaces for global land areas. Int J Climatol. 2017;37(12):4302–15. doi:10.1002/joc.5086.
- Batjes NH, Ribeiro E, Van Oostrum A. Standardised soil profile data to support global mapping and modelling (WoSIS snapshot 2019). Earth Sys Sci Data. 2020;12(1):299–320. doi:10.5194/essd-12-299-2020.
- Guerrero J, Andrello M, Burgarella C, Manel S. Soil environment is a key driver of adaptation in Medicago truncatula: new insights from landscape genomics. New Phytologist. 2018;219(1):378–90. doi:10.1111/nph.15171.
- Lasky JR, Forester BR, Reimherr M. Coherent synthesis of genomic associations with phenotypes and home environments. Mol Ecol Resour. 2018;18(1):91–106. doi:10.1111/1755-0998.12714.
- Rathinam M, Mishra P, Mahato AK, Singh NK, Rao U, Sreevathsa R. Comparative transcriptome analyses provide novel insights into the differential response of Pigeonpea (Cajanus cajan L.) and its wild relative (Cajanus platycarpus (Benth.) Maesen) to herbivory by Helicoverpa armigera (Hübner). Plant Mol Biol. 2019;101(1):163–82. doi:10.1007/s11103-019-00899-7.
- Yu F, Liang K, Fang T, Zhao H, Han X, Cai M, Qiu F. A group VII ethylene response factor gene, ZmEREB180, coordinates waterlogging tolerance in maize seedlings. Plant Biotechnol J. 2019 Dec;17(12):2286–98.
- Prescott-Allen C, Prescott-Allen R. The first resource: wild species in the North-American economy. New Haven: Yale University Press; 1986.
- Brar DS, Khush GS. Alien introgression in rice. Oryza: from molecule to plant. Plant Molecular Biology. 1997;35:35–47.
- Rick CM, Chetelat RT. editors. Utilization of related wild species for tomato improvement. Leuven, Belgium; 1995.
- Crute IR. From breeding to cloning (and back again?): a case study with lettuce downy mildew. Annu Rev Phytopathol. 1992;30(1):485–506. doi:10.1146/annurev.py.30.090192.002413.
- Pink DAC, Walkey DGA. Resistance in marrow (Cucurbita pepo L.) to different strains of cucumber mosaic virus. J Agric Sci. 1984;103(3):519–21. doi:10.1017/S0021859600043616.
- Glaszmann J-C, Kilian B, Upadhyaya HD, Varshney RK. Accessing genetic diversity for crop improvement. Curr Opin Plant Biol. 2010;13(2):167–73. doi:10.1016/j.pbi.2010.01.004.
- Pandey MK, Upadhyaya HD, Rathore A, Vadez V, Sheshshayee MS, Sriswathi M, Govil M, Kumar A, Gowda MVC, Sharma S, et al. Genomewide association studies for 50 agronomic traits in peanut using the ‘reference set’comprising 300 genotypes from 48 countries of the semi-arid tropics of the world. PLoS one. 2014;9(8):e105228. doi:10.1371/journal.pone.0105228.
- Caniato FF, Hamblin MT, Guimaraes CT, Zhang Z, Schaffert RE, Kochian LV, Magalhaes JV. Association mapping provides insights into the origin and the fine structure of the sorghum aluminum tolerance locus. AltSB PLoS One. 2014;9(1):e87438. doi:10.1371/journal.pone.0087438.
- Carvalho JG, Schaffert RE, Malosetti M, Viana JHM, Menezes CB, Silva LA, Guimaraes CT, Coelho AM, Kochian LV, van Eeuwijk FA, et al. Back to acid soil fields: the citrate transporter SbMATE is a major asset for sustainable grain yield for sorghum cultivated on acid soils. G3: Genes, Genomes, Genetics. 2016;6(2):475–84. doi:10.1534/g3.115.025791.
- Pocnet C, Dupuis M, Congard A, Jopp D. Personality and its links to quality of life: mediating effects of emotion regulation and self-efficacy beliefs. Motiv Emot. 2017;41(2):196–208. doi:10.1007/s11031-017-9603-0.
- Leonelli S, Davey RP, Arnaud E, Parry G, Bastow R. Data management and best practice for plant science. Nat Plants. 2017;3(6):1–4. doi:10.1038/nplants.2017.86.