ABSTRACT
Epidemiological characteristics and molecular features of carbapenem-resistant Enterobacter (CR-Ent) species remain unclear in China. In this study, we performed a genomic study on 92 isolates from Enterobacter-caused infections from a multicenter study in China. Whole genome sequencing (WGS) was used to determine the genome sequence of 92 non-duplicated CR-Ent strains collected from multiple tertiary health centres. The precise species of Enterobacter strains were identified by average nucleotide identity (ANI) and in silico DNA–DNA hybridization (isDDH). Molecular features of high-risk CR-Ent sequence type (ST) lineages and carbapenemase-encoding plasmids were determined. The result revealed that the most common human-source CR-Ent species in China was E. xiangfangensis (66/92, 71.93%), and the proportion of carbapenemase-producing Enterobacter (CP-Ent) in CR-Ent was high (72/92, 78.26%) in comparison to other global regions. Furthermore, ST171 and ST116 E. xiangfangensis were the major lineages of CP-Ent strains, and ST171 E. xiangfangensis was more likely to cause infections in older patients. Genomic analysis also highlighted the likelihood of intra-hospital/inter-hospital clonal transmission of ST171 and ST116 E. xiangfangensis. In addition, the blaNDM-harbouring IncX3-type plasmid was identified as the prevalent carbapenemase-encoding plasmid carried by CR-Ent strains, and was experimentally confirmed to be able to self-transfer with high frequency. This study detailed the genomic and clinical characteristics of CR-Ent in China in the form of multicenter for the first time. The high risk of carbapenemase-producing ST171 and ST116 E. xiangfangensis, and the blaNDM-harbouring IncX3-type plasmid were detected and emphasized.
The genus Enterobacter is the third most common pathogenic Enterobacteriaceae species in human after Escherichia and Klebsiella, which can cause a variety of clinical infections, including bloodstream and intra-abdominal infections [Citation1,Citation2]. Carbapenems are the main drugs for the treatment of multidrug-resistant Enterobacter infections, but the rise of carbapenem resistance in Enterobacter spp., primarily caused by the acquisition of carbapenemase-encoding genes (blaKPC, blaNDM, blaVIM, etc.), has significantly compromised the choice of clinical antimicrobial treatment [Citation3,Citation4].
Identification of Enterobacter is still a challenging process. Although automated microbial identification systems and mass spectrometers are reliable for bacterial species identification, they are not specific enough for detecting Enterobacter species [Citation1]. Clinical laboratories usually only classify Enterobacter species as E. cloacae complex, resulting in the inability to correlate antimicrobial resistance to Enterobacter at the species level. The combination of average nucleotide identity (ANI) and in silico DNA–DNA hybridization (isDDH) based on whole-genome sequencing analysis was recently recommended for more precise identification of Enterobacter species [Citation5]. These methods provide high resolution to further divide Enterobacter into different species.
Carbapenemase-encoding genes have been increasingly detected in Enterobacter strains. Recently, Zong et al. examined 4899 high-quality Enterobacter genome sequences within GenBank and carried out the precise species identification and preliminary resistance gene screening [Citation3]. The result showed that E. xiangfangensis was the most common species globally, and blaNDM gene was mostly encountered in China. Several high-risk Enterobacter clones associated with carbapenem resistance have also been described. The carbapenemase-producing ST171 E. xiangfangensis has been recognized as a high-risk lineage in the USA [Citation6,Citation7]. Additionally, ST90, ST93 and ST114 E. xiangfangensis, and ST78 E. hoffmannii are also globally distributed and are associated with multiple carbapenemases (VIM, NDM, KPC, and OXA-48) [Citation8]. However, our understanding of precise species and genomic features of clinical carbapenem-resistant Enterobacter (CR-Ent) in China is still limited.
Herein, we collected 92 non-duplicated CR-Ent strains causing clinical infections from nine hospitals in China. Whole genome sequencing (WGS) and genomic analysis were conducted to characterize the precise species and genomic features of these strains.
Materials and methods
Bacterial strains
The multicenter study covered nine tertiary health care grade hospitals from Beijing, Chongqing, Guangdong, Jiangsu, Ningxia, Sichuan, and Yunnan. These provinces or municipalities are located in North China (Beijing), East China (Jiangsu), South China (Guangdong), Southwest China (Chongqing, Sichuan, and Yunnan), and Northwest China (Ningxia), respectively. All archived carbapenem-resistant Enterobacterales (CRE) strains from nosocomial infections were collected from the above nine hospitals between January 2017 and March 2021 (data not shown), and 92 CR-Ent strains were further selected (Table S1). Detailed bacterial strain collection was described in the Supplemental Text 1. Clinical data were obtained from medical chart using the same standardized questionnaire in each hospital.
All high-quality Enterobacter genome sequences within GenBank database were acquired for further analysis (details in Supplemental Text 1).
Sequencing and assembly
Bacterial genomic DNA of 92 screened CR-Ent strains was isolated using the Omega Bio-Tek Bacterial DNA Kit (Doraville, GA, USA). Further WGS and assembly were performed (details in Supplemental Text 1).
Identification of bacterial species
Precise species identification was performed by calculating the pairwise isDDH and ANI value between the genome sequence of the query strain and those of type strains of Enterobacter spp. described recently [Citation3] (details in Supplemental Text 1).
Multi-locus sequence typing (MLST) analysis and phylogenetic analysis
The sequence types (STs) of Enterobacter strains were identified according to the online MLST scheme (https://pubmlst.org/). Further phylogenetic analysis was performed on the ST116 and ST171 E. xiangfangensis strains (details in Supplemental Text 1).
Sequence annotation and comparison
Open reading frames (ORFs) and pseudogenes were predicted using RAST 2.0 [Citation9] combined with BLASTP/BLASTN searches against the UniProtKB/Swiss-Prot database [Citation10] and the RefSeq database [Citation11]. Annotation of resistance genes, virulence genes, mobile elements, and other features was carried out using the online databases, including ResFinder 4.1 [Citation12], VFDB (core dataset) [Citation13], ISfinder [Citation14], and Tn Number Registry [Citation15]. Multiple and pairwise sequence comparisons were performed using BLASTN.
Conjugal transfer
A conjugal transfer experiment was carried out using sodium azide-resistant E. coli J53 as the recipient and the E. xiangfangensis strains HD7405, HD8705, and HD2956 as the donor (details in Supplemental Text 1).
Growth curve analysis
Strains were grown overnight in 3 ml of LB with shaking (200 rpm) at 37°C, and were then diluted to an OD600 of 0.25. Next, 2 μl of the solution was added to 200 μl LB in a 96-well plate in triplicate. Culture densities were determined every 10 min by measuring the OD600 for 12 h with shaking (200 rpm) at 37 °C via FLUOstar Omega (BMG Labtech, Germany). Growth curves were estimated via GraphPad Prism 5.0 (GraphPad Software, Inc.) using a two-way analysis of variance (ANOVA), and p-value < 0.05 was considered to be statistically significant.
Statistical analysis
Unpaired two-tailed Student’s t-tests or Mann–Whitney U test were performed to analyse the statistical significance of measurement data, while the chi-square test or Fisher exact test were performed to analyse the statistical significance of categorical data. All data were analysed using GraphPad Prism 5.0 (GraphPad Software, Inc.), and p-value <0.05 was considered to be statistically significant.
Nucleotide sequence accession numbers
The complete sequences of chromosome and plasmids from HD8642 were submitted to GenBank under BioProject PRJNA824643, while the sequences of the rest 91 Enterobacter strains were submitted to GenBank under BioProject PRJNA824645. In addition, the sequences of 28 mapped plasmids were submitted to GenBank under accession number ON209123-ON209150.
Results
Identification and STs of Enterobacter strains
A total of 92 nonduplicated CR-Ent strains were identified in this study (Table S1), including 64 from Chengdu, 13 from Yinchuan, 6 from Suzhou, 5 from Chongqing, 3 from Kunming and 1 from Guangzhou. Most of these strains were initially reported as Enterobacter cloacae or E. cloacae complex in clinical laboratories. Precise species identification revealed that the most common CR-Ent species was E. xiangfangensis (n = 66), while the next common Enterobacter species was E. hoffmannii (n = 16) (). The remaining strains were classified into six Enterobacter species: E. chengduensis (n = 4), E. mori (n = 2), E. hormaechei (n = 1), E. kobei (n = 1), E. roggenkampii (n = 1), and E. chuandaensis (n = 1). Additionally, E. xiangfangensis was also the most prevalent Enterobacter species in each region where CR-Ent was collected (66.67−100%).
Table 1. Proportion of Enterobacter species and STs of carbapenem-resistant Enterobacter strains sequenced in this study.
These 92 Enterobacter strains belong to 38 STs (including six unnamed novel STs), and the most prevalent lineages were ST116 (n = 15) and ST171 (n = 14) E. xiangfangensis. Except for Chengdu and Yinchuan, the distribution of CR-Ent was very diverse, without a predominant clone. The most prevalent lineages in Chengdu were ST171 (12/64, 18.75%) and ST116 (8/64, 12.50%) E. xiangfangensis, while ST116 (5/13, 38.41%) in Yinchuan.
Carbapenemase-producing Enterobacter (CP-Ent) strains
Most of the 92 CR-Ent strains carried carbapenemase-encoding genes (72/92, 78.26%) (), which included 71 carrying a sole carbapenemase-encoding gene (blaNDM-1 [n = 45], blaNDM-5 [n = 18], blaIMP-26 [n = 5], blaIMP-4 [n = 2] and blaVIM-1 [n = 1]) and one harbouring two genes (blaNDM-1 and blaVIM-1). The blaNDM was the most prevalent carbapenemase-encoding gene in each region (66.67−100%), except Guangzhou.
According to the method described in the method section, all high-quality Enterobacter genome sequences within GenBank database were acquired (dated 4 June 2021). We further determined 1402 human-source Enterobacter genome sequences carrying carbapenemase-encoding genes and having geographic information (Table S2). These genomes and the 72 genomes sequenced in this study were further analysed. The results showed that globally E. xiangfangensis remained to be the most common CP-Ent species from human sources (Table S3) and the prevalence of CP-E. xiangfangensis was high (>37%) in Asia, Africa, Europe, Oceania, and North or South America. CP-E. hoffmannii, E. cloacae, and E. asburiae were also commonly detected, but their prevalence was lower than that of E. xiangfangensis.
Nevertheless, the prevalent CP-Ent lineages in different regions vary significantly (Table S3). The result showed that ST171 E. xiangfangensis was the most common in Asia (including China) and North America, while ST114 E. xiangfangensis was common in Europe, ST90 E. xiangfangensis in Oceania, ST510 E. xiangfangensis in South America, and ST78 E. hoffmannii and ST456 E. cloacae in Africa. Additionally, some lineages were only prevalent in one region, such as ST116 E. xiangfangensis in China and ST830 E. xiangfangensis in Oceania, the prevalence rates of which were second in their respective regions.
A total of 1505 carbapenemase-encoding genes were identified in the above 1474 genome sequences (Table S4). For class A carbapenemase-encoding genes, the blaKPC was most common in South America (mainly blaKPC-2) and North America (mainly blaKPC-3), while the blaIMI (mainly blaIMI-1) was most common in the Asia (except China). Among class B carbapenemase-encoding genes, the blaNDM was prevalent in Asia, Africa, and Europe (mainly blaNDM-1). Besides blaNDM-1, blaNDM-5 was also frequently detected in China. In addition, blaIMP was mostly encountered in Asia (mainly blaIMP-1) and Oceania (mainly blaIMP-4), and blaVIM was typically detected in Europe (mainly blaVIM-1). For class D carbapenemase-encoding genes, blaOXA-48-like gene was most common in Asia and Europe (mainly blaOXA-48).
In general, the most common CP-Ent species from human sources in the world was E. xiangfangensis, while the specific prevalent Enterobacter lineages and carbapenemase-encoding genes showed large geographical variations. Since ST171 and ST116 E. xiangfangensis were the most prevalent lineages in China, we then performed a detailed genetic dissection analysis on the genome sequences of the two lineages.
Phylogenetic analysis of ST171 and ST116 strains
The STs of all high-quality Enterobacter genome sequences within GenBank database described above were further identified. We detected 264 human-source ST171 E. xiangfangensis genome sequences and 41 human-source ST116 E. xiangfangensis genome sequences, which had geographic information, including both carbapenemase gene producers and non-producers (Table S5). Phylogenetic analysis based on core SNPs was performed on 278 ST171 E. xiangfangensis genome sequences [including 14 sequenced in this study, and 264 from GenBank], and 56 ST116 E. xiangfangensis strains [including 15 sequenced in this study, and 41 from GenBank], respectively.
The phylogenetic tree of ST171 E. xiangfangensis strains revealed that 278 ST171 E. xiangfangensis strains were divided into four major separately clustering clades (). Most of the ST171 E. xiangfangensis strains carried carbapenemase-encoding genes (265/278, 95.32%), which included 264 carrying a sole carbapenemase-encoding gene (blaKPC-3 [n = 161], blaNDM-1 [n = 53], blaKPC-4 [n = 21], blaNDM-5 [n = 16], blaKPC-2 [n = 6], blaOXA-48 [n = 4], blaVIM-1 [n = 2] and blaVIM-4 [n = 1]) and one harbouring two genes (blaKPC-4 and blaIMP-13). The blaKPC-carrying ST171 E. xiangfangensis strains were the most common (189/278, 67.99%). Among these strains, blaKPC-3-carrying strains were mainly classified into clade IV (160/161, 99.38%) and mainly distributed in the USA (160/161, 99.38%), while all blaKPC-4-carrying ones (including the strain co-harbouring blaKPC-4 and blaIMP-13) were all classified into clade II and were evenly distributed in the USA (11/22, 50.00%) and the UK (11/22, 50.00%). Furthermore, most blaNDM-carrying ST171 E. xiangfangensis strains were classified into clade III (68/69, 98.56%) and mainly distributed in Asia (62/69, 89.86%). These results showed that blaNDM-carrying ST171 E. xiangfangensis most likely originated and was predominant in Asia and had a tendency to spread to other continents. Additionally, the strains carrying blaKPC-2, blaOXA-48, blaVIM-1, blaVIM-4, or blaIMP-13 had no obvious geographical distributions due to the small quantity.
Figure 1. A maximum-likelihood phylogenetic tree of ST171 E. xiangfangensis strains. A total of 278 ST171 E. xiangfangensis strains (including 14 sequenced in this study, and 264 from GenBank) were performed with phylogenetic analysis, while E. cloacae strain ATCC 13047 (GenBank accession number NC_014121) was used as the out group. Bar corresponds to scale of sequence divergence.
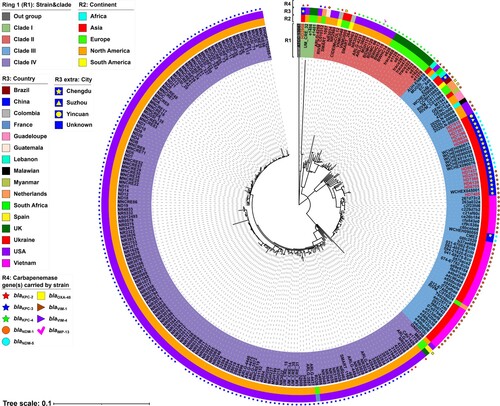
The core SNPs of 74 ST171 E. xiangfangensis strains from clade III were further pairwise compared to determine their clonal relatedness (Table S6). In the 22 ST171 strains from China, there were 0–112 SNPs difference, suggesting that they did not belong to a single strain. However, the sporadic clonal transmission of ST171 strains was still detected; for instance, HD7296, HD8634, and HD8606 had 0–1 SNP difference and were collected from different patients in the same ward in a hospital in Chengdu. Additionally, the inter-hospital clonal transmission of ST171 strains was also detected. For example, WCHEH090011, WCHEH090023, WCHEH090059, and WCHEX016162 were collected from another hospital in Chengdu [Citation5] and shared 2–4 SNPs difference with HD8671 and HD7437 from our study, supporting the inter-hospital transmission of NDM producing ST171 strains in China.
For ST116 E. xiangfangensis, 56 strains were divided into two major separately clustering clades (). Among them, only 17 strains carried carbapenemase-encoding genes, including 8 blaNDM-1, 7 blaNDM-5, 1 blaKPC-4, and 1 blaIMP-4. Of note, all 15 blaNDM-carrying strains were from China and were evenly distributed in the two clades (). These results showed that blaNDM-carrying ST116 E. xiangfangensis was also an important lineage in China, and the carbapenem resistance in this ST was driven by the independent acquisition of the blaNDM gene in two major phylogenetic clades.
Figure 2. A maximum-likelihood phylogenetic tree of ST116 E. xiangfangensis strains. A total of 56 ST171 E. xiangfangensis strains (including 15 sequenced in this study, and 41 from GenBank) were performed with phylogenetic analysis, while E. cloacae strain ATCC 13047 (GenBank accession number NC_014121) was used as the out group. Bar corresponds to scale of sequence divergence.
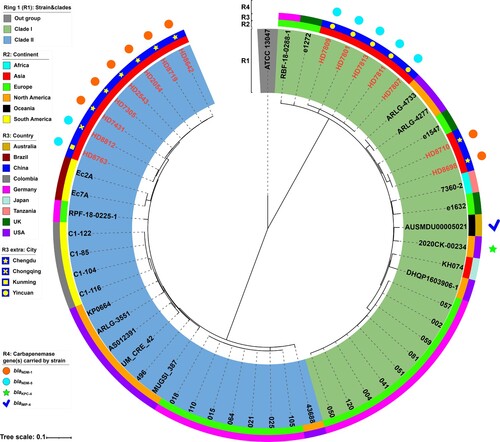
The core SNPs of 56 ST116 E. xiangfangensis strains were also pairwise compared (Table S7). In the 15 ST116 strains from China, there were 1–3604 SNPs difference. Similarly, the regional clonal transmission of ST116 strains was found in Chengdu and Yinchuan. For example, HD7801, HD7807, HD7809, HD7811, and HD7813 had 1–5 SNPs difference and were collected from different patients in the same ward in a hospital in Yinchuan, while HD7305 and HD7431 had 2 SNPs difference, and were from different patients in the same ward in a hospital in Chengdu.
Clinical and genetic features of different Enterobacter groups
The clinical data of patients with CR-Ent were collected (Table S1). Further statistical analysis was conducted between ST116 E. xiangfangensis (n = 15), ST171 E. xiangfangensis (n = 14), and the other CR-Ent strains (n = 63), including age, sex, isolation source, time to positive culture, community or hospital onset, admission route, length of hospital stay, post-culture length of hospital stay, and 30-day outcome of patient after positive culture (). The results showed that compared with patients with the other CR-Ent strains, patients with ST171 E. xiangfangensis strain were older (p < 0.05), while other factors did not significantly differ among groups. However, the proportion of poorer 30-day outcomes (worsened/died/withdrawing treatment) of patients with ST116 (6/15, 40.00%) and ST171 (6/14, 42.86%) E. xiangfangensis strains after positive culture was relatively higher compared with patients with the other CR-Ent strains (16/63, 25.40), respectively.
Table 2. Statistical analysis of Enterobacter strains sequenced in this study.
The virulence genes and acquired antimicrobial resistance genes carried by 92 CR-Ent strains were identified through VFDB (core dataset) [Citation13] and ResFinder 4.1 [Citation12] (Table S8 and S9). Further statistical analysis was also conducted between three groups (). Genes that were statistically different among groups were further listed. For virulence genes, the result showed that compared with the other CR-Ent strains, ST116 E. xiangfangensis strain carried significantly more virulence genes (p < 0.001) while ST171 E. xiangfangensis strain carried fewer virulence genes (p < 0.05). ST116 E. xiangfangensis strain was more likely to carry iroDCBNE, csgG, cgsD, vipA/tssB, vipB/tssC, hcp/tssD, tssF, and acrA genes (p < 0.05). Additionally, though ST171 E. xiangfangensis strain carried fewer virulence genes, it was more likely to carry entE, fepD, cgsD, vipB/tssC, and ugd genes (p < 0.05). For antimicrobial resistance genes, the number of antimicrobial resistance genes carried by ST116 E. xiangfangensis strain and ST171 E. xiangfangensis strain had no significant difference compared with the other CR-Ent strain. However, ST116 E. xiangfangensis strain and ST171 E. xiangfangensis strain were more likely to carry blaNDM gene (p < 0.05) and were less likely to carry mcr-9 gene (p < 0.05) compared with the other CR-Ent strains.
Growth curves of ST171 and ST116 strains
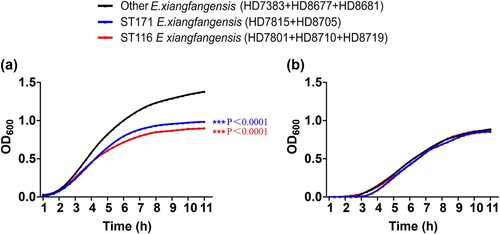
The growth assay was performed to compare the proliferative capability of ST171 and ST116 E. xiangfangensis with other ST carbapenem-resistant E. xiangfangensis. The 15 ST116 E. xiangfangensis strains sequenced in this study were divided into three groups according to the phylogenetic analysis (Figure S1a), while 14 ST171 E. xiangfangensis strains sequenced in this study were divided into two groups (Figure S1b). One randomly selected strain from each of the five groups, and three randomly selected strains from other STs of E. xiangfangensis strains, were included. The growth of ST116 E. xiangfangensis and ST171 E. xiangfangensis was slower (p < 0.001) in LB without antibiotic, as evidenced by decreases in optical density compared with the optical densities of other ST E. xiangfangensis, respectively (a). However, the growth curves of ST116 E. xiangfangensis and ST171 E. xiangfangensis showed no significant difference (p > 0.05) with other ST carbapenem-resistant E. xiangfangensis strains in LB with 2 μg/mL meropenem (b). The results showed that the fitness cost of ST171 and ST116 E. xiangfangensis was not obvious under carbapenem selection pressure.
Plasmid analysis of ST171 and ST116 strains
The complete sequences of carbapenemase-encoding plasmids carried by 14 ST171 E. xiangfangensis strains and 15 ST116 E. xiangfangensis strains sequenced in this study were obtained. Fifteen carried blaNDM-5, 13 carried blaNDM-1, and one carried blaVIM-1 (Table S10). All blaNDM-5 genes (n = 15) were carried by IncX3-type plasmid, while blaNDM-1 genes were carried by IncX3-type plasmid (n = 12) or IncFII-type plasmid (n = 1) (a). Further analysis revealed that all blaNDM genes were carried by Tn125-related derivatives (). Additionally, the blaVIM-1 gene was located within the integron In916 and carried by an unknown-type plasmid pHD2956-VIM (b), which was identical to the reference plasmid pCY-VIM (GenBank accession number KF998104) [Citation16]. The detailed genetic analysis of plasmids and Tn125-related derivatives is described in Supplemental Text 2.
Figure 4. Linear comparison of carbapenemase-encoding plasmids from ST171 and ST116 E. xiangfangensis strains. Genes are denoted by arrows. Genes, mobile genetic elements and other features are coloured based on function classification. Shading regions denote homology of two plasmids (light blue: ≥ 99% nucleotide identity). The names of plasmids from ST171 E. xiangfangensis strains were marked in red, while ST116 E. xiangfangensis strains in blue. The names of reference plasmids were marked in black. The accession number of plasmids pNDM5_090011, pNDM-HN380, pNDM1_045001 and pCY-VIM used as reference are CP036312, JX104760, CP043383 and KF998104, respectively.
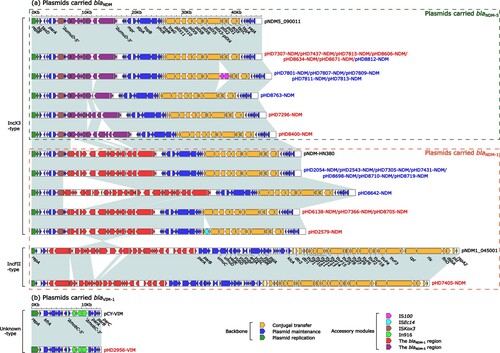
Figure 5. Organization of the blaNDM regions from blaNDM-harbouring plasmids. Genes are denoted by arrows. Genes, mobile genetic elements and other features are coloured based on their functional classification. Shading denotes regions of homology (light blue: ≥ 99% nucleotide identity). The accession number of Tn125 used as reference is JN872328 [Citation20].
![Figure 5. Organization of the blaNDM regions from blaNDM-harbouring plasmids. Genes are denoted by arrows. Genes, mobile genetic elements and other features are coloured based on their functional classification. Shading denotes regions of homology (light blue: ≥ 99% nucleotide identity). The accession number of Tn125 used as reference is JN872328 [Citation20].](/cms/asset/97b025f1-e831-4694-983c-6de3934410bf/temi_a_2148562_f0005_oc.jpg)
Besides plasmids from ST171 and ST116 strains, the carbapenemase-encoding plasmids carried by other ST Enterobacter strains sequenced in this study were also preliminarily analysed. The result showed that blaNDM-harbouring IncX3-type plasmids were the most common ones (n = 51) in all carbapenemase-encoding plasmids, while other plasmid types only accounted for a small proportion, such as blaNDM-harbouring IncC and IncFII plasmids (data not shown). Additionally, all blaNDM genes were carried by Tn125-related derivatives.
The IncFII-type plasmid pHD7405-NDM and IncX3-type plasmid pHD8705-NDM could be transferred into E. coli J53 from strain HD7405 and HD8705 through conjugal transfer experiment, respectively. However, repeated conjugation attempts failed to transfer pHD2956-VIM from strain HD2956 into E. coli J53, which is likely due to the lack of tra module in this plasmid (b).
To assess the impact of IncFII-type and IncX3-type plasmids on the growth of strain, the growth of E. coli J53 transconjugants of IncFII-type plasmid pHD7405-NDM or IncX3-type plasmid pHD8705-NDM was compared. The growth curves of E. coli J53 and J53/pHD8705-NDM (IncX3) showed no significant difference (p > 0.05), while the growth of J53/pHD7405-NDM (IncFII) was clearly slower (p < 0.05) than the parental E. coli J53 (Figure S2). The results indicated that the acquisition of IncX3-type plasmid had minimal impact on the growth of the parental strain, but the acquisition of IncFII-type plasmid had a high fitness cost in vitro growth.
Discussion
CR-Ent has become a new emerging threat to global public health [Citation4]. However, poor identification of Enterobacter species further complicates the analysis of CR-Ent species. This study used ANI and isDDH to examine 92 non-duplicated CR-Ent strains from Chinese patients. Most of these strains were initially reported as Enterobacter cloacae or E. cloacae complex in clinical laboratories. Nevertheless, the precise species identification showed that the two most common CR-Ent species were E. xiangfangensis and E. hoffmanni, while E. cloacae was not detected. Furthermore, due to the main mechanism for carbapenem resistance in Enterobacter was the producing carbapenemase, we screened the carbapenemase-encoding genes in the strains from this study and publicly available genomes. The result showed that the most common CP-Ent species from human sources in the world was E. xiangfangensis, while the specific prevalent Enterobacter lineage and carbapenemase-encoding genes vary in different regions. Additionally, compared with 24% CP-Ent prevalence (46/192) in the USA [Citation17], the proportion of CP-Ent in CR-Ent was much higher (78.26%, 72/92) in China (p < 0.05). It indicated distinct molecular mechanisms underlying the carbapenem resistance in Enterobacter between China and the USA, and the acquisition of carbapenemase genes and plasmids has been identified as the main cause of carbapenem resistance in China. Our results also highlighted the geographical impacts of rapid molecular diagnosis on CR-Ent. Molecular carbapenemase gene detection can likely capture ∼80% CR-Ent in China and only some ∼25% in the USA.
CP-ST171 E. xiangfangensis has been recognized as a high-risk lineage in the USA [Citation6–8], which could cause infections in healthcare settings, and the clonal spread of ST171 CR-Ent has also been documented. Unlike the previously reported CP-ST171 E. xiangfangensis that mainly carried blaKPC, the strains in China mainly carried blaNDM. Indeed, the same feature was also found in CP-ST171 E. xiangfangensis from several other Asian countries. Phylogenetic analysis further revealed that all blaNDM-carrying ST171 E. xiangfangensis from Asia were from the same clade, suggesting that this lineage most likely originated in Asia and evolved from the same clone. Additionally, CP-ST116 E. xiangfangensis was also identified as an important lineage in China. CP-ST116 E. xiangfangensis was rarely reported in previous studies, and only two CP-ST116 E. xiangfangensis were deposited in the GenBank database. However, among the CP-strains sequenced in this study, ST116 E. xiangfangensis (20.83%, n = 15) showed the highest prevalence, exceeding ST171 E. xiangfangensis (19.44%, n = 14). These ST116 E. xiangfangensis carried blaNDM, and although belonging to two different clades, they are indicative of independent acquisition of blaNDM by the different ST116 strains in China. Of note, further analysis highlighted the intra-hospital clonal transmission of ST116 and ST171 E. xiangfangensis and the inter-hospital clonal transmission of ST171 E. xiangfangensis. The inter-hospital clonal transmission of ST171 E. xiangfangensis was also recently reported in a bloodstream infection-associated study in China [Citation5]. Although statistical analysis of clinical data showed that ST171 and ST116 E. xiangfangensis were not significantly associated with poorer outcomes, ST171 E. xiangfangensis was more likely to infect older patients. However, the number of Enterobacter strains collected in this study was relatively limited; thus, more studies are needed to evaluate these findings.
Horizontal gene transfer is one of the main carbapenemase genes transfer mechanisms. In this study, blaNDM-harbouring IncX3-type plasmids were the most common plasmids among all carbapenemase-encoding plasmids. The IncX3-type plasmid was self-transmissible, which has been confirmed in this study and previous reports [Citation18,Citation19]. Furthermore, the same IncX3-type plasmid was detected in E. xiangfangensis strains belonging to different clones. For example, HD7307 and HD8634 were collected from patients in different wards of the same hospital and belonged to two ST171 clones (56 SNPs between each other). The interval between the collection dates of the two strains was about one month. However, pHD7307-NDM from HD7307 was identical to pHD8634-NDM from HD8634. The self-transferability of blaNDM-harbouring IncX3-type plasmid in part explained the wide-ranging existence of this plasmid in Enterobacter strains. Additionally, all blaNDM genes from strains sequenced in this study were carried by Tn125-related derivatives. Tn125 was a composite transposon flanked by two copies of ISAba125 [Citation20]. Tn125 is considered important in the initial dissemination of blaNDM to plasmid backbones [Citation21]. In general, self-transmissible blaNDM-harbouring plasmids should be closely monitored in China, especially blaNDM-harbouring IncX3-type plasmids widely distributed in Enterobacter strains.
The reasons for the high prevalence of ST116 and ST171 E. xiangfangensis in China need to be further explored. However, the result of statistical analysis showed that ST171 and ST116 E. xiangfangensis were more likely to carry some important virulence genes or antimicrobial resistance genes, such as iroDCBNE (involving the encoding of siderophore salmochelin) in ST116 E. xiangfangensis, and cgsD (involving the encoding of curli fibres), vipB/tssC (involving the encoding of T6SS), and blaNDM in ST116 and ST171 E. xiangfangensis. The presence of these genes may provide certain selection advantages against host immune factors or hospital environments. IncX3-type plasmids were reported as the most common ones carrying blaNDM, which were mainly distributed in China and neighbouring countries [Citation22]. Our results showed that the fitness cost of carrying an IncX3-type plasmid was minimal compared to other plasmids. Furthermore, compared with the major blaKPC-harbouring IncFIA-type plasmid carried by the ST171 strain in the USA [Citation7,Citation23], the IncX3-type plasmid was adapted to carriage by enterobacteria that colonize mammalian hosts [Citation24]. Therefore, the ability of strains to acquire and maintain the blaNDM-harbouring IncX3-type plasmid might be one major driver in the prevalence of ST171 and ST116 E. xiangfangensis in China.
This study has some limitations. Firstly, the sample size of CR-Ent in this study was relatively small. The small sample size was in part because we focused on isolates associated with clinical infections and only non-duplicated isolates were included. However, this study was one of the largest studies focusing on both clinical and genomic features of CR-Ent in China, and the isolates were collected from different geographical regions, and the predominant CR-Ent clones and plasmids were described. Secondly, although all high-quality Enterobacter genome sequences within GenBank were included in this study, the Genbank genomes may be biased towards submission sources with genome sequencing capacity. Nevertheless, this dataset represents the most comprehensive CR-Ent genomic resources currently available and the data provide important distribution information and genomic features of different global CR-Ent strains.
In conclusion, the most common CR-Ent strains recovered from humans in China were not E. cloacae but E. xiangfangensis, which is consistent with the finding that most CP-Ent strains recovered from human sources (including China) were E. xiangfangensis. The results also suggested that some species-specific factors may contribute to the predominance of E. xiangfangensis in CR-Ent, which deserves further studies. In China, ST171 and ST116 E. xiangfangensis were the major clinical lineages of CP-Ent strains and were mainly associated with blaNDM-harbouring IncX3-type plasmids. Further surveillance and infection control measures should be advocated to prevent the clonal dissemination of CP-ST171 and ST116 E. xiangfangensis, and the horizontal dissemination of blaNDM-harbouring IncX3-type plasmids.
Supplemental Material
Download Zip (2.4 MB)Data availability statement
The data presented in the study are included in the article/supplementary files, further inquiries can be directed to the corresponding author/s.
Acknowledgments
The authors are grateful for professor Dongsheng Zhou from State Key Laboratory of Pathogen and Biosecurity, Beijing Institute of Microbiology and Epidemiology for the assistance in the drawing of figures.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Mezzatesta ML, Gona F, Stefani S. Enterobacter cloacae complex: clinical impact and emerging antibiotic resistance. Future Microbiol. 2012 Jul;7(7):887–902.
- Davin-Regli A, Lavigne JP, Pagès JM. Enterobacter spp.: update on taxonomy, clinical aspects, and emerging antimicrobial resistance. Clin Microbiol Rev. 2019 Sep 18;32(4):e00002–19.
- Zong Z, Feng Y, McNally A. Carbapenem and colistin resistance in enterobacter: determinants and clones. Trends Microbiol. 2021 Jun;29(6):473–476.
- Nordmann P, Poirel L. Epidemiology and diagnostics of carbapenem resistance in gram-negative bacteria. Clin Infect Dis. 2019 Nov 13;69(Suppl 7):S521–S528.
- Wu W, Wei L, Feng Y, et al. Precise species identification by whole-genome sequencing of enterobacter bloodstream infection, china. Emerging Infect Dis. 2021 Jan;27(1):161–169.
- Hawken SE, Washer LL, Williams CL, et al. Genomic investigation of a putative endoscope-associated carbapenem-resistant enterobacter cloacae outbreak reveals a wide diversity of circulating strains and resistance mutations. Clin Infect Dis. 2018 Jan 18;66(3):460–463.
- Gomez-Simmonds A, Annavajhala MK, Wang Z, et al. Genomic and geographic context for the evolution of high-risk Carbapenem-resistant Enterobacter cloacae complex clones ST171 and ST78. mBio. 2018 May 29;9(3):e00542–18.
- Peirano G, Matsumura Y, Adams MD, et al. Genomic epidemiology of global Carbapenemase-Producing Enterobacter spp., 2008-2014. Emerging Infect Dis. 2018 Jun;24(6):1010–1019.
- Brettin T, Davis JJ, Disz T, et al. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep. 2015 Feb 10;5:8365.
- Boutet E, Lieberherr D, Tognolli M, et al. Uniprotkb/Swiss-Prot, the manually annotated section of the UniProt knowledge base: how to use the entry view. Methods Mol Biol. 2016;1374:23–54.
- O'Leary NA, Wright MW, Brister JR, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016 Jan 4;44(D1):D733–D745.
- Zankari E, Hasman H, Cosentino S, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012 Nov;67(11):2640–2644.
- Liu B, Zheng D, Zhou S, et al. VFDB 2022: a general classification scheme for bacterial virulence factors. Nucleic Acids Res. 2022 Jan 7;50(D1):D912–D917.
- Siguier P, Perochon J, Lestrade L, et al. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006 Jan 1;34(Database issue):D32–D36.
- Tansirichaiya S, Rahman MA, Roberts AP. The transposon registry. Mob DNA. 2019;10:40.
- Yang L, Wu AW, Su DH, et al. Resistome analysis of Enterobacter cloacae CY01, an extensively drug-resistant strain producing VIM-1 metallo-β-lactamase from China. Antimicrob Agents Chemother. 2014 Oct;58(10):6328-6330.
- van Duin D, Arias CA, Komarow L, et al. Molecular and clinical epidemiology of carbapenem-resistant Enterobacterales in the USA (CRACKLE-2): a prospective cohort study. Lancet Infect Dis. 2020 Jun;20(6):731–741.
- Xiao X, Zeng F, Li R, et al. Subinhibitory concentration of colistin promotes the conjugation frequencies of mcr-1- and blaNDM-5-positive plasmids. Microbiol Spectr. 2022 Mar 1:e0216021.
- Chen R, Liu Z, Xu P, et al. Deciphering the epidemiological characteristics and molecular features of blaKPC-2- or blaNDM-1-positive Klebsiella pneumoniae isolates in a newly established hospital. Front Microbiol. 2021;12:741093.
- Poirel L, Bonnin RA, Boulanger A, et al. Tn125-related acquisition of blaNDM-like genes in Acinetobacter baumannii. Antimicrob Agents Chemother. 2012 Feb;56(2):1087–1089.
- Acman M, Wang R, van Dorp L, et al. Role of mobile genetic elements in the global dissemination of the carbapenem resistance gene blaNDM. Nat Commun. 2022 Mar 3;13(1):1131.
- Wu W, Feng Y, Tang G, et al. Ndm metallo-β-lactamases and their bacterial producers in health care settings. Clin Microbiol Rev. 2019 Mar 20;32(2)::e00115–18.
- Pereira EC, Anacker M, Houseman J, et al. A cluster of carbapenemase-producing Enterobacter cloacae complex ST171 at a tertiary care center demonstrating an ongoing regional threat. Am J Infect Control. 2019 Jul;47(7):767–772.
- Baomo L, Lili S, Moran RA, et al. Temperature-regulated IncX3 plasmid characteristics and the role of plasmid-encoded H-NS in thermoregulation. Front Microbiol. 2021;12:765492.