ABSTRACT
Zika virus (ZIKV) infections are typically asymptomatic but cause severe neurological complications (e.g. Guillain–Barré syndrome in adults, and microcephaly in newborns). There are currently no specific therapy or vaccine options available to prevent ZIKV infections. Temporal gene expression profiles of ZIKV-infected human brain microvascular endothelial cells (HBMECs) were used in this study to identify genes essential for viral replication. These genes were then used to identify novel anti-ZIKV agents and validated in publicly available data and functional wet-lab experiments. Here, we found that ZIKV effectively evaded activation of immune response-related genes and completely reprogrammed cellular transcriptional architectures. Knockdown of genes, which gradually upregulated during viral infection but showed distinct expression patterns between ZIKV- and mock infection, discovered novel proviral and antiviral factors. One-third of the 74 drugs found through signature-based drug repositioning and cross-reference with the Drug Gene Interaction Database (DGIdb) were known anti-ZIKV agents. In cellular assays, two promising antiviral candidates (Luminespib/NVP-AUY922, L-161982) were found to reduce viral replication without causing cell toxicity. Overall, our time-series transcriptome-based methods offer a novel and feasible strategy for antiviral drug discovery. Our strategies, which combine conventional and data-driven analysis, can be extended for other pathogens causing pandemics in the future.
Introduction
ZIKV has emerged as a serious public health concern due to mild clinical symptoms (e.g. fever, arthralgia, skin rash) and severe neurological manifestations (e.g. neonatal microcephaly and Guillain–Barré syndrome in adults) [Citation1]. Due to the limited knowledge of the molecular mechanisms by which ZIKV counteracts the host immune response and causes neuropathogenesis, there is currently neither a licensed vaccine for ZIKV nor an effective treatment for the virus. It is imperative to understand the pathogenic mechanisms and how it exploits the host’s immune response.
Targeting host factors rather than viral enzymes has emerged as a viable therapeutic approach to suppress viral replication [Citation2]. Understanding the dynamic response of host molecules during viral infection is necessary for the development of better therapeutic strategies. High temporal-resolution transcriptomes have emerged as an attractive technology to capture the detailed dynamics of virus-host interactions at genome-wide gene-expression levels, and to identify pro-/anti-viral host factors during virus infection in an unbiased and comprehensive strategy [Citation3]. Recently, by systematically and thoroughly assessing host gene expression changes in naturally ZIKV-infected myeloid dendritic cells, Xiaoming et al. found that ZIKV infection resulted in profound downregulation of interferon-stimulated genes (ISGs) and upregulation of a large number of ZIKV dependency genes [Citation4]. Conventional drug discovery is a laborious, time-consuming process with low success rates and high costs [Citation5]. Drug repurposing has gained considerable attention due to its potential in repositioning known drugs to treat emerging diseases [Citation6]. Signature-based drug repositioning is a technique that scores negative correlations between drugs and diseases using gene expression profiles. This approach assumes that a drug with the ability to reverse transcriptome alterations in host cells caused by viral infections is a possible antiviral [Citation6]. The connectivity map (CMAP) database and Library of Integrated Network-based Cellular Signatures (LINCS) project have contributed significantly to the rapid development of drug repurposing by freely providing the scientific community with a large number of gene signatures of drugs or compounds [Citation7–9]. By embedding expression data of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) into CMAP data for signature matching, Belyaeva A et al. identified serine/threonine and tyrosine kinases as possible drug targets for the therapy of COVID-19 [Citation10]. Large-scale drug-target databases (e.g. ChEMBL, DrugBank) are publicly available. Access to such databases will facilitate drug repurposing.
Here, by performing a time-series transcriptome (public data: GSE98889) analysis of infected HBMECs ranging from 3 h post-infection (hpi) to 216 hpi (more detail in the original publications [Citation11]), we found that ZIKV effectively evaded activation of immune responses, completely reprogrammed the cellular transcriptome, and identified genes might critical for ZIKV infection. To further explore the roles of these genes in viral replication, loss-of-function experiments were conducted in vitro. What's more, data from a set of independent clinical trials were interrogated to estimate their expression patterns during ZIKV infection. Finally, signature-based drug repositioning and cross-reference with drug-target databases revealed known as well as novel potential drugs/compounds and further evaluated the antiviral activity of these drugs at the cellular level.
Materials and methods
Affymetrix gene array analysis
Affymetrix CEL files were downloaded from the NCBI Gene Expression Omnibus (GEO GSE98889 [Citation11]). Using the R Bioconductor packages oligo, Limma, and Mfuzz, the raw intensities were subjected to RMA background correction, summarization, log2-transformation, differential expression analysis, and clustering.
RNA-seq data set and analysis
A set of 11 RNAseq samples derived from fetal membrane cells of three ZIKV-positive pregnant women, who were infected with ZIKV at different trimesters in Singapore in 2016, and two healthy females (for more information, see the original publication and BioProject accession number PRJNA578707 [Citation12]) were downloaded from NCBI SRA. The RNAseq analysis pipeline nf-core/rnaseq [Citation13] was used to map the raw RNA-seq data to the human genome (GRCh38) using the following parameters: nextflow run nf-core/rnaseq –input samplesheet.csv –fasta Genome.fa –gtf GtfFile.gtf –star_index STAT_directory. The count reads matrix was normalized using variance stabilizing transformation in DESeq2. The RNAseq normalized expression matrix was used to confirm microarray expression analysis.
Functional enrichment analysis
Gene Ontology (GO) and Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathway enrichment analysis were performed on gene sets from https://biit.cs.ut.ee/gprofiler using clusterProfiler 4.0 and metascape.
Co-expression and different co-expression analysis
Time-series gene expression under mock and ZIKV infection was analyzed using Short Time-series Expression Miner (STEM) with default parameters to identify gene sets showing significantly different temporal expression patterns, respectively [Citation14]. TimesVector [Citation15] was performed to identify gene sets showing similar and different gene expression patterns between mock and ZIKV infections with parameters “TimesVector -g Matrix_expression.txt -c 2 -t 5 -d m -k 200 -o Prefix”.
Collection of ZIKV-dependent and immune-related genes
ZIKV-dependent genes were identified based on genome-wide RNAi and CRISPR/Cas9 screens [Citation16]. Immune-related genes came from GO term “GO:0002376 immune system process”, “GO:0031047 gene silencing by RNA”, all immune-related KEGG pathways (KEGG id: 03050, 04062, 04215, 04610, 04611, 04612, 04620, 04621, 04622, 04623, 04625, 04640, 04650, 04657, 04658, 04659, 04660, 04061, 04662, 04664, 04666, 04670, 04672, 05160) in https://www.genome.jp/kegg/pathway.html, and the two immunity databases InnateDB and ImmPort (https://www.immport.org/).
Nichenet analysis
To better understand the regulatory mechanisms driving the co-expression patterns of TVC7, the ligand–receptor, signalling and gene regulatory networks deposited in the NicheNet database (https://zenodo.org/record/3260758) were explored using the R package nichenetr according to the tutorial in https://workflows.omnipathdb.org/nichenet1.html. Network graphs were produced using Graphviz (http://www.graphviz.org) with Kdot as the layout algorithm.
Drug repurposing
To identify potential antivirals against ZIKV, two different approaches, signature-based drug repositioning and cross-reference with drug-target databases, were used. The function queryDGIdb in R package rDGIdb was used to implement cross-reference of genes in TVC7 with DGIdb. Due to agents capable of reversing virus-induced changes in gene expression profiles were more promising antivirals, R package signatureSearch was used to score gene expression signature based on CMAP method and LINCS database [Citation8], with genes in TVC7 and TVC48 as inputs.
Cheminformatics analysis
The R packages webchem, ChemmineR, and openbabel were used to obtain the chemical structures and biochemical descriptions of antiviral candidates. Chemical similarity analysis was performed for 74 drugs using the get.fingerprint function in R package rcdk. R packages ChemmineR, fingerprint, and hclust were used to calculate the Tanimoto similarity score and performed hierarchical clustering.
Virus, cells, and reagents
Vero cells (African green monkey kidney epithelial cells) (CCL-81, American Type Culture Collection) were cultured in minimum essential medium (MEM) (Life Technologies) supplemented with 10% fetal bovine serum (FBS) (Gibco, NY, USA) at 37 °C and 5% CO2. Human brain endothelial cells (HBMECs, passage 6–18), provided by Prof. Cui Min (State Key Laboratory of Agricultural Microbiology, College of Veterinary Medicine, Huazhong Agricultural University, Wuhan, China), were cultured in Roswell Park Memorial Institute 1640 medium (RPMI-1640) (Life Technologies) supplemented 10% FBS (Gibco) at 37 °C and 5% CO2. ZIKV (Zika virus/SZ01/2016/China, GenBank: KU866423.2) was obtained from the Wuhan Institute of Virology, Chinese Academy of Science.
RNA isolation and real-time quantitative PCR (RT-qPCR)
RNA was isolated from cells or supernatants using TRIzol Reagent (Invitrogen) according to the manufacturer's instructions. First-strand cDNA was synthesized using FastKing RT Kit (TIANGEN, China) according to the manufacturer's instructions. For each reaction, the quantitative PCR (qPCR) mix (20 μL) consisted of cDNA derived from 50 ng RNA, forward primer, reverse primer, and iTap™ Universal SYBR Green® Supermix (Bio-Rad). RT-qPCR was performed using Bio-Rad CFX real-time quantitative PCR system with the preprogramme: activation at 95°C for 3 min, 40 cycles of 95°C for 5 s and 60°C for 45s. All primers for RT-qPCR are listed in Table S1. Transcript levels and viral RNA copies were quantified using the relative standard curve method. Gene expression data were analyzed using the threshold cycle (Ct) and 2 −ΔΔ Ct method. To evaluate the copy number of ZIKV in the samples, the RT-qPCR threshold cycle value was set according to the standard curve obtained with plasmids containing DNA sequences amplified from the corresponding ZIKV cDNA at serial dilutions ranging from 1 × 103 to 1 × 108 copies/mL.
Plaque assay
1× 105 Vero cells were seeded in 24-well plates in DMEM (10% FBS) and incubated with serially diluted samples (10−1 to 10−6 in serum-free DMEM) for 2 h at 37°C, 5% CO2. After incubation, the medium was replaced with semi-solid medium comprising DMEM supplemented with 1% (w/v) carboxymethylcellulose (Sigma) and 2% (v/v) FBS. After incubation for 4 days, the semi-solid medium was gently removed and cells were fixed with 4% paraformaldehyde (PFA) (25 °C, 20 min) and stained with crystal violet solution (1% crystal violet, 50% methanol, 37 °C, 1 h). Viral lysis plaques were counted and viral titres were expressed as plaque-forming units per millilitre of medium (PFU/mL).
Viability assays
HBMECs were seeded in 96-well microplates at a density of 104 cells per well 1 d under 37°C, 5% CO2. The cells were treated with serial dilutions of the drug in RPMI-1640 supplemented with 10% FBS, in triplicate. After 48 h of incubation at 37°C, 5% CO2, the medium was replaced with fresh RPMI-1640 with 10% FBS supplemented with 10 μL Cell Counting Kit-8 (CCK8) reagent (Sigma). After incubation for 3 h at 37°C, 5% CO2, the absorbance of each well was measured at 450 nm with the Fluostar Omega Plate Reader (BMG Labtech).
RNA interference
HBMECs were seeded in 24-well plates and transfected with 50 nM double-stranded siRNA oligonucleotides, which were designed and synthesized by RiboBio (Guangzhou, China), according to the manufacturer's instructions 24 h prior to infection with ZIKV at MOI 0.1. Then, cells were recovered and examined for viral RNA by RT-qPCR. Non-targeting siRNA (NT siRNA) was used as a control. Triplicates were performed for each treatment. No cellular toxicity was observed for either siRNA or transfection agent. The efficiency of knockdown was determined by qPCR. Sequences of siRNA against specific targets were listed in Table S2.
Drug treatment
HBMECs were seeded in 24-well plates and infected with ZIKV (MOI = 1) in serum-free RPMI-1640 at 37°C, 5% CO2 for 2 h, then the medium was replaced with RPMI-1640 supplemented with 10% FBS and serially diluted drugs (dissolved in DMSO, Table S3). After 48 hpi, viral RNA in supernatants was extracted using a Viral RNA Mini Kit (Qiagen), according to the manufacturer’s protocol. Viral RNA copies were examined by RT-qPCR. Viral titre in supernatants was quantified by plaque assay.
Statistical analysis
The statistical software R 4.1.0 and GraphPad Prism v.7 were used to analyze the data. Statistical significance was assessed by unpaired student t-test. Multiple comparisons were performed by one-way analysis of variance. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001. Dose–response curves and CC50 values were calculated using a log-logistic regression (“LL.2”) with a four-parameter logistic curve in R package drm.
Results
Less pronounced expression changes in ZIKV-infected HBMECs
Previous evidence indicated that the ability of ZIKV to efficiently infect HBMECs could damage the central nervous system (CNS) and cause neurological disease [Citation11]. To gain a comprehensive understanding of the cellular response to ZIKV, we used time-series transcriptome (GSE98889) data to explore the dynamic of the host response to viral infection. Transcriptome data were obtained by microarrays for HBMECs collected at 12, 24, 48, 72, and 216 hpi [Citation11]. Firstly, principal component analysis (PCA) was performed on the normalized matrix to assess the biological variability among samples ((a)). As a result, the first principal component (PC1) captured most of the variation between samples attributable to variations across time points, which explained 36% of the total variance, while PC2 explained significant differences between mock and ZIKV-infected samples.
Figure 1. changes in the global gene expression of HBMECs infected by ZIKV. (a) Principal-component analysis (PCA) of mock- and ZIKV-infected samples in GSE98889. Expression values were log-transformed and scale-normalized. (b) Numbers of genes with significant differential expression (|log 2 (foldchange)| > 1 & P value < 0.05), orange and blue barplot indicate upregulated and downregulated genes, respectively. (c) Venn diagram depicting the overlap of significant DEGs between different time points. (d) Enriched gene ontology (GO) terms associated with “viral” and “virus” at each time point. (e) Time-course expression profiles of 941 differential expression genes across treatment. Columns indicate separate arrays and rows indicate genes grouped by Mfuzz clusters. Then the panel represents time-series differential expression (log 2 (foldchange)), the distribution of differential expression values across the time course (boxplot), the cumulative differential activity (barplot), and the direction of differential expression (heatmap, orange and blue indicate upregulation and downregulation).
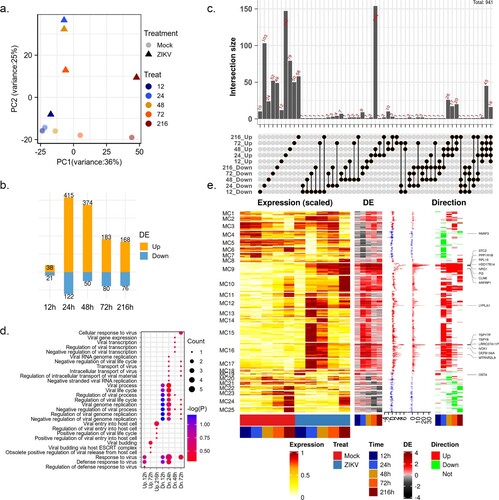
After viral infection, global changes in gene expression were rather modest. A total of 941 differentially expressed genes (DEGs, P value < 0.05 and |log2 FC| > 1, (b–c)) were identified. The number of DEGs peaked at 24 hpi with 537 genes and then decreased at 48 and 72 hpi. ((b)). In the original report, the number of infected cells, ZIKV titres, and viral RNA levels peaked at 48 hpi and then declined [Citation11]. These suggested that viral replication in HBMECs peaked after an extensive host transcriptional response triggered by ZIKV infection. More than 60% of DEGs were time-specific and a considerable number of DEGs (mainly up-regulated genes) shared at 24 and 48 hpi ((c)). Among upregulated genes, 126 genes were induced in at least three-time points tested post-infection, 16 (ARFRP1, BOD1, DHX16, HSD17B14, LRRC37A11P, LYPLA1, MTRNR2L9, NOX1, NRG1, PI3, PPP1R1B, RPL19, SPTLC3, STC2, TSPY7P, and TSPY8) of which were consistently upregulated. Frustratingly, no functional terms were significantly enriched on these genes. Among Gene Ontology (GO) terms related to the virus life cycle, “defense response to virus” and “negative regulation of viral genome replication” were upregulated at the early stage of viral infection but downregulated at the late stage ((d)).
To better understand the dynamics of the intracellular environment after ZIKV infection, 941 DEGs were clustered into 25 clusters using the Mfuzz algorithm ((e)). The most pronounced changes in gene expression during ZIKV infection occurred mainly at 24 hpi ((e)). The differential expression dynamics of most individual genes are clearly pulsatile and transient changes (e.g. MC5-7 and MC10-18). Genes in MC9 and MC25 showed sustained activation and repression, respectively ((a)). These genes might play important roles in viral infection. In cluster MC9 ((e)), RPL19 was required for the replication of yellow fever virus (YFV), West Nile virus WNV and ZIKV [Citation17]. Further functional characterization of these genes to confirm their roles in ZIKV infection was warranted.
ZIKV infections evaded the activation of immune-related genes
To further investigate how ZIKV altered the intracellular environment to conduct infection, we analyzed the expression pattern of ZIKV-dependency genes, which were screened by genome-wide CRISPR/Cas9 and small interfering RNA (siRNA) using the H1-HeLa cell line and the ZIKV MR766 strain [Citation4,Citation16]. As expected, a large proportion (44%) of ZIKV-dependency genes were up-regulated during infection. 36 out of these genes were consistently up-regulated during the infection time course ((a)). Of note, these transcripts contained several proteins involved in viral replication, such as ADP-ribosylation factor-related protein 1 (ARFRP1) and ceramide synthase 5 (CERS5). ARFRP1 interacted with NS5A, and silencing of ARFRP1 dramatically restrained HCV replication in both subgenomic replicon cells and HCVcc-infected cells [Citation18]. Ceramide, a bioactive sphingolipid associated with signalling and apoptosis, was required for ZIKV infection and closely associated with ZIKV NS4B [Citation19]. These genes were enriched in GO terms such as “maintenance of Golgi location”, “septin cytoskeleton organization”, “B cell activation involved in immune response”, and “positive regulation of T cell-mediated immune response to tumour cell” ((b)).
Figure 2. Expression of ZIKV-dependency genes and immune-related genes. Time-course expression profiles of ZIKV-dependency genes (a) and immune-related genes (d). (b) Representative GO terms for DEGs related to ZIKV-dependent genes. (c) The distribution of differential expression values of DEGs related to the immune system. (e) Representative GO terms for DEGs related to the immune system.
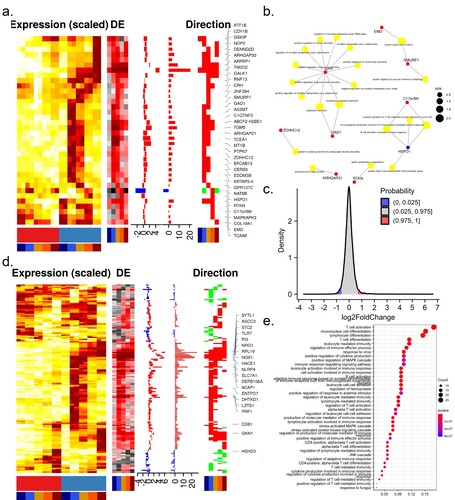
Understanding the contribution of immune-related genes to viral infections will shed light on the pathogenesis of diseases and aid in the therapeutic development. To this end, the expression pattern of immune-related genes was investigated. More than 95% of immune genes were dysregulated at very low levels during the infection time course ((c)), suggesting that ZIKV might be able to evade the host immune response by avoiding triggering it. 168 immune genes were significantly dysregulated at least one sampling time during the infection time course.16 out of these genes (ACP6, ASCC3, CLDN23, FABP5, FCAMR, IL1F10, IL36A, KLF4, NBL1, NRG1, PI3, RPL19, RPS27A, STC2, SYTL1, TLR7) presented in cluster MC9 and were consistently up-regulated, 2 genes (TOX, WDR62) presented in cluster MC25 and were consistently downregulated; others were transiently dysregulated ((d)). GO analysis revealed that 168 immune genes mainly involved in response to the virus, JNK cascade, T cell and lymphocyte-mediated immunity ((e)), which might cause progressive T-cell dysfunction, compromising viral clearance and resulting in persistent infection. Among these genes, TLR7 could effectively trigger NF-κB activation, cytokine secretion, and the inflammatory response; IL36A and IL1F10 were cytokines that effectively activated pro-inflammatory response. These genes could confer potential resistance to ZIKV.
Additionally, six immune genes (MAPKAPK3, PTPN7, HSPD1, C17orf99, GMFB, IFIT1B) were also ZIKV-dependency genes. Over-expression of MAPKAPK3 hindered INF-α-induced gene expression and inhibited IFN-α-mediated antiviral activity [Citation20].
ZIKV rewired host transcriptional structures
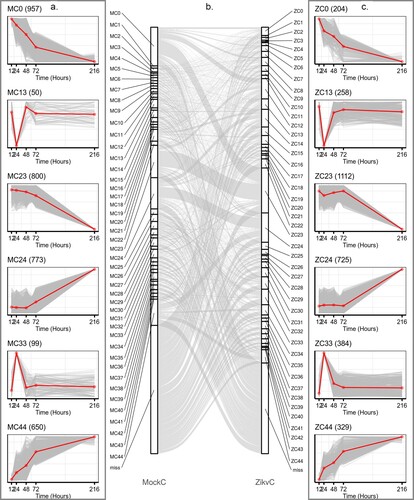
We hypothesis that ZIKV could redirect host cells’ transcriptional machinery so that viral genes would be expressed more effectively. Identification of the differences between mock and ZIKV-infected transcriptional environments would be useful to reveal gene sets critical for viral replication. To this end, 8305 and 7376 variable genes in the time course of mock and ZIKV infections were identified using Short Time-series Expression Miner (STEM) software [Citation14], and categorized into 45 distinct temporal expression clusters, respectively (, Figure S1, S2 & Table S4). The algorithms implemented in STEM were deliberately developed for clustering short time-series data [Citation14]. Although the temporal expression patterns were similar during mock and ZIKV infections (Figure S1–S2), the composition of cluster members with similar expression patterns differed significantly under the two conditions, e.g. MC0 vs ZC0, MC44 vs ZC44 (). This suggested that ZIKV infection reprogrammed the host gene expression. The cumulative expression patterns in ZC0 and ZC44 were negatively and positively correlated with viral RNA accumulation, respectively (). To gain a biological understanding of genes in these two clusters, GO enrichment-functional analyses were performed with Metascape. Cluster ZC0, composing 204 genes, mainly enriched in terms involved in “adaptive immune system”, “negative regulation of leukocyte migration”, “negative regulation of immune system process”, and “detection of biotic stimulus”; Cluster ZC44, composing 329 genes, mainly enriched in terms involved in “interleukin-36 pathway”, “TNFR1-induced NF-κB signalling pathway” and “apoptotic cleavage of cellular proteins” (Figure S3).
Figure 3. ZIKV infection reprogrammed host transcriptional structures. Representative gene clusters were identified by STEM clustering under mock- (a) and ZIKV (c) infection conditions, respectively. Red lines indicate the mean expression level of genes in each cluster. The number in the upper right corner of each panel represents the number of genes in each cluster. (b) Sankey diagram showing gene expression patterns of transition between mock (left side)- and ZIKV- infected (right side) conditions.
Additionally, TimesVector was a triclustering algorithm developed specifically for three-dimensional data (i.e. gene-time-condition) to capture gene expression patterns that were similar or different between two conditions [Citation15]. Using TimesVector, 7769 variable genes were identified and assigned to 63 different co-expression clusters (Figure S4 & Table S4). These clusters could be broadly segregated into 2 main modules: gene expression patterns under mock infection were negatively correlated with those under viral infection (e.g. TVC48 and TVC127, Figure S4); the gene expression patterns were altered only at specific stages of infection (e.g. TVC44 and TVC135, Figure S4). Once again, these results suggested that ZIKV infection reprogrammed the host gene expression.
Prioritizing genes in module TVC7 and testing their effect on viral infection
By combining the results of the Mfuzz algorithm and TimesVector, we found that genes in TCV7 and TVC163 were consistently activated and showed distinct co-expression patterns over the time course of mock and ZIKV infection, respectively ((a–b)). Genes in TVC7 showed a higher level of differential expression pattern during the time course of ZIKV infection compared to genes in TVC163 which showed a moderate differential expression pattern (Figure S4). Therefore, genes in TVC7 might be most closely associated with the progression of ZIKV infection. Subsequently, we investigated the genes in TVC7 and found that activation of some genes (e.g. ASCC3, BAG1, KLF4, NRG1, TLR7, SYTL4) could suppress host immunity and enhance viral infection [Citation21–26].
Figure 4. Prioritizing genes in module TVC7. (a) Venn diagram depicting the overlap between Mfuzz cluster MC9, and different co-expression modules TVC7 and TVC163. TVC7 and TVC163 are two clusters containing a set of genes, calculated by TimesVector v1.5. (a) vectorized clustering approach to the analysis of time series transcriptome data from two or more sample conditions, https://github.com/inukj/TimesVector). (b) TVC7 showed distinct co-expression patterns throughout the time course in response to mock and ZIKV infection. (c) The interactions of TVC7 proteins with viral proteins in HVIDB. (d) The regulatory network of TVC7. Each node indicated a protein, and each regulatory edge represented regulatory relationships predicted by NicheNet or host-virus protein-protein interactions in HVIDB. (e) Validation of the gene expression in TVC7 after ZIKV infection at 24 hpi using qPCR. (f) Validation of the gene expression in TVC7 in an external, clinical, and independent dataset from PRJNA578707. (g) Validation of genes in TVC7 via siRNA-mediated knockdown. ZIKV infection relative to NT siRNA control following siRNA-mediated knockdown of genes in TVC7 measured by the expression level of viral RNA, MV represents mock treatment before virus infection.
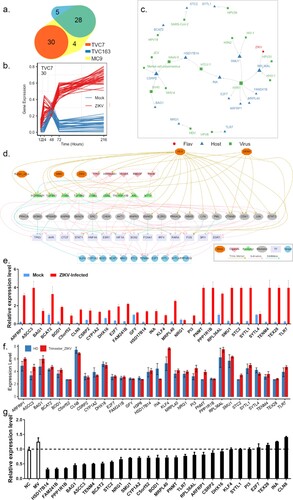
By cross-referencing the genes in TVC7 with Human Viral Interaction Database (HVIDB), a comprehensive database of 48,643 experimentally validated human-viral PPI [Citation27], we found that half (15) of genes in TVC7 could interact with viral proteins, RPL36AL could interact with the polyprotein of ZIKV ((c)). RPL36AL also interacted with NS1, a nonstructural protein of H1N1, US11, a cofactor of HHV-1, and E2, a regulatory protein of HPV31. RPL36AL encoded a ribosomal protein that might involve in the translation of viral mRNA.
Genes in TVC7 showed close co-expression patterns during ZIKV infection and might share common regulators (e.g. ligands, receptors, transcription factor, TFs). To infer ligand–receptor–TFs interactions that derived the expression patterns of TVC7, we used the R package OmnipathR to access https://omnipathdb.org/ and capture inter- and intracellular protein interactions during signalling, transcriptional and post-transcriptional regulation. Among the predicted ligands, the tumour protein TP53 had a strong regulatory potential for several genes in TVC7, such as ASCC3, E2F7, NRG1, TENM4, KLF4 and STC2 (Figure S5A–S5C). TP53 was downregulated at 48 hpi, but upregulated at late stage of viral infection. In previous studies, TP53 was considered to be a key component within the regulatory network related to ZIKV infection and neonatal microcephaly [Citation28]. Moreover, the predicted ligands HSP90AA1, P4HB, HBEGF and S100A4 exhibited similar gene expression accumulation to TP53 in the settings of viral infection. Among these 16 predicted ligands, the over-expression of S100A4 facilitated ZIKV entry into macrophages and persistence in varicose vasculature [Citation29]. In HVIDB, 10 out of these predicted ligands directly interacted with viral proteins. Notably, TP53, HSP90AA1, P4HB and MST1 could interact with many viral proteins of flaviviruses (e.g. HCV and DENV, Figure S5G–S5H).
OmnipathR predicted 91 receptors in signalling downstream of 16 ligands mentioned above and mediated the expression of genes in TVC7 (Figure S5D–S5F). In HVIDB, 46 out of these receptors interacted with viral proteins. HSP90AB1, TNFRSF10B, ITGB1, CD82, TGFBR1, TGFBR2, ITGB3, and MTTP, directly interacted with proteins of flaviviruses (e.g. DENV, HCV, WNV and Kunjin virus). HSP90AB1, TNFRSF10B and CD82 were regulated by TP53; TGFBR1, TGFBR2 and ITGB3 were regulated by HSP90AA1; ITGB1 and ITGB3 were regulated by VEGFA; MTTP was regulated by P4HB (Figure S5D). AXL was a ZIKV entry factor and mediated persistent ZIKV infection [Citation30]. Here, we found that AXL was predicted to be a receptor for VEGFA.
Based on ligand–receptor–TFs regulatory networks from https://omnipathdb.org/ and flaviviruses-host protein–protein interactions from HVIDB, we further reconstructed the TVC7 gene regulatory network ((d)). Due to the different weights assigned to each interaction by Omnipath, the final regulatory network consisted of 5 ligands, 9 receptors, 19 signalling transducers, 14 TFs and 11 genes in TVC7 ((d)). In this regulatory network, more than half of genes were involved in immune system regulation such as MAPK3, MAPK8, LYN, IKBKB, CHUK, IRF4, SRC, STAT1, STAT3; 24 proteins interacted with flaviviruses. These results suggested that ZIKV might evade host immunity and promote virus replication by interacting with multiple regulators of TVC7.
Validation of genes in TVC7
The expression of genes in (exclude H3P6, a pseudogene) TVC7 was verified at 24 hpi by qPCR analysis. Most of these genes were upregulated in HBMECs after viral infection, whereas GFY and SYTL4 showed no significant changes ((e)). The expression of these genes was also validated in an external, clinical, and independent dataset of trimester-specific full-term placentas from three ZIKV-infected and two healthy women ((f)) [Citation12]. Then, loss-of-function screens were performed to assess their role in viral infection. After HBMECs were transfected with control or specific siRNAs for 24 h, the expression levels of these genes decreased, thus confirming the validity of siRNAs. Of the three individual siRNAs, the one with the best silencing efficiency was used for subsequent experiments (Table S2). After transfection with siRNA for 24 h, HBMECs were infected with ZIKV and viral RNA level was examined at 24 hpi. As shown in (g), RNAi knockdown of these genes in HBMECs severely affected viral replication.
Prediction and validation of anti-ZIKV agents
The above experiments showed that genes in TVC7 were required for ZIKV infection. Then drugs targeting these genes would be able to block ZIKV infection. To validate this notion, we cross-referenced the genes in TVC7 with DGIdb, which contained over 100,000 curated drug-gene interactions [Citation31]. If our analysis was indeed of clinical pertinence, then drugs with known efficacy in reducing ZIKV infection should be identified. We found that CYP1A2, E2F7, KLF4, NRG1, PI3, PPP1R1B and TLR7 were targeted by 611 drugs (Table S3). Of these drugs, 26 (amphotericin, baicalein, chloroquine, chlorpromazine, clomipramine, cyclosporin, cytarabine, didanosine, floxuridine, fluphenazine, gemcitabine, hydroxychloroquine, hydroxyzine, imiquimod, indinavir, menadione, N-(3-chlorophenyl)-6,7-dimethoxyquinazolin-4-amine, nevirapine, niclosamide, promethazine, pyridostigmine, simeprevir, stavudine, triflupromazine, wortmannin, zidovudine) were retrieved from the PubChem database as potential antiviral agents. 12 compounds (baicalein, chloroquine, chlorpromazine, cyclosporin, floxuridine, gemcitabine, hydroxychloroquine, hydroxyzine, imiquimod, niclosamide, promethazine, simeprevir) were reported as anti-ZIKV agents [Citation32–41]. Drugs that affect the activity of multiple host factors are promising antiviral candidates. Among the nine drugs targeting more than one gene ((a)), six (tamoxifen, aspirin, chloroquine, colchicine, erlotinib, and genistein) inhibit ZIKV replication in vitro and in vivo [Citation33,Citation42–46]. Fluoxetine suppressed DENV at an early stage of viral replication [Citation47]. These results suggested that the genes in TVC7 might be candidate targets for the development of anti-ZIKV agents, other drugs or compounds identified here might be potential anti-ZIKV agents.
Figure 5. Predicted anti-ZIKV agents and their effect on viral infection. (a) Candidate drugs that target TVC7. Drug candidates against TVC7 were identified by cross-referencing with DGIdb. Drugs targeted more than two genes in TVC7 or retrieved from the PubChem database as potential antiviral agents were shown. Red fonts indicated known anti-ZIKV agents. (b) Chemical structure similarity clustering of drugs identified by signature-based drug repositioning (top 40) and cross-reference with DGIdb. Different coloured branches indicated compounds with distinct chemical structures. Genes with orange colours represent known anti-ZIKV agents, darkred represents compounds being tested in this work, and grey represents others identified here. (c) and (d), ZIKV infectious titres and RNA levels in the culture supernatant of HBMECs at 48 h post-infection with ZIKV at MOI 1 and treated with the indicated compounds, 20 μM Ribavirin, or equivalent volumes of the solvents DMSO. DMSO served as a negative control for all compounds. ZIKV RNA levels were measured by RT-qPCR. ZIKV titres were measured by plaque assay. Data were analysed with student’s t-test, (*p < 0.05; **p < 0.01, ***p < 0.001, ****p < 0.001). Bars represent the mean ± SD of three biological replicates.
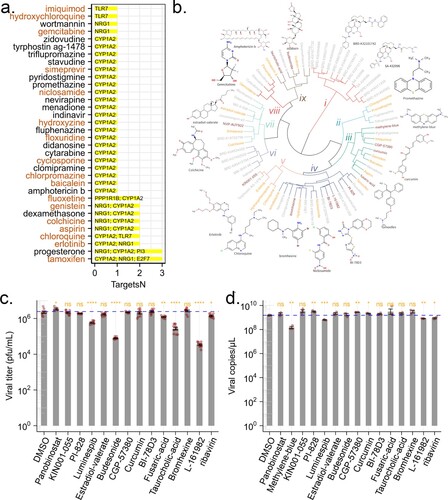
In signature-based drug repositioning, gene expression profiles were used to compare drug-disease correlations. By using the CMAP method in signatureSearch V1.6.1, and LINCS database [Citation8], drugs or compounds capable of reversing gene expression patterns of TVC7 and TVC48 (which were up- and down-regulated, respectively, throughout the time course of infection) were predicted. Of the top 40 agents with predicted activity, three (amodiaquine, estradiol-valerate and ouabain) were reported as anti-ZIKV agents [Citation48–50], methylene-blue and curcumin were thought to interact directly with viral proteins or viral capsid proteins to inhibit viral replication [Citation51,Citation52]. Here, methylene-blue and curcumin here might be host-directed therapeutic agents.
Drugs sharing similar chemical structures might share common biological activities, but might lead to cross-intolerance. To find novel anti-ZIKV agents, the chemical structures of the above-identified drugs or compounds were obtained from PubChem through ChemmineR, and were grouped into 9 most representative chemical clusters based on their fingerprints through R package rcdk and hclust ((b)). Compounds with anti-ZIKV activity were present in clusters ii, iii, iv, v, vii, viii, and ix ((b)). Therefore, we suggested that other drugs or compounds in clusters ii, iii, iv, v, vii, viii, and ix might be anti-ZIKV agents. To verify this hypothesis, the compounds (Panobinostat, methylene-blue, KIN001-055, PI-828, Luminespib/NVP-AUY922, estradiol-valerate, budesonide, CGP-57380, curcumin, BI-78D3, fusaric-acid, taurocholic acid, bromhexine, L-161982) which shared similar chemical structures to known anti-ZIKV agents ((b)) were selected and evaluated the antiviral activity at the cellular level. Firstly, the highest non-toxic compound concentrations and half maximal cytotoxic concentration 50 (CC50) of each compound were determined (Table S3) by the cell counting kit 8 (CCK8) assay. We infected HBMECs with ZIKV at an MOI of 1 and treated them with each compound, an equimolar concentration of the solvent (DMSO) or ribavirin as a positive control. Out of the 14 compounds, treatment with methylene-blue, Luminespib/NVP-AUY922, budesonide, fusaric-acid, taurocholic acid, L-161982 reduced the viral titres. Two of these compounds, budesonide and L-161982 induced a > 100-fold reduction in ZIKV infectious titres. Methylene-blue completely abrogated production of infectious progeny viruses. In addition to the plaque-based assay, we also tested for inhibition using an RT-qPCR assay. Treatment with methylene-blue, Luminespib/NVP-AUY922 and L-161982 resulted in reduction of intracellular ZIKV RNA levels.
Discussion
ZIKV infection causes congenital defects (e.g. microcephaly) and severe neurological disease manifestations (e.g. Guillain–Barré syndrome). However, there are no therapeutic drugs or effective vaccines available to treat ZIKV infection. In this study, we combined temporal gene expression analysis, bioinformatics, drug assay databases, and cheminformatics with wet-lab experiments to get a list of candidate drugs that could be repositioned as antivirals. Here, we found that ZIKV effectively circumvented the dysregulation of immune response genes and completely reprogrammed the cellular transcriptome; siRNA-mediated knockdown validated the genes, which were progressively upregulated during viral infection but showed distinct expression patterns between ZIKV- and mock infection, and revealed novel proviral and antiviral factors (e.g. CYP1A2, PPP1R1B and TLR7). Signature-based drug repositioning and cross-reference with DGIdb, identified 74 drug candidates, one-third of which were known anti-ZIKV agents (e.g. chloroquine, hydroxychloroquine); Two promising antiviral candidates (Luminespib/NVP-AUY922 and L-161982) were validated in cellular assays to reduce viral replication without inducing cytotoxicity. Overall, our approaches based on time-series transcriptome provide a novel and viable strategy for antiviral drug discovery. Our strategies which combine conventional and data-driven analysis can be extended for other pathogens causing pandemics in the future.
Unlike bacteria, viruses must replicate in the cellular environment. For survival, viruses have developed diverse strategies to rearrange host metabolic, transcriptional and signalling pathways to create an environment favourable for virus amplification and spread. RNA viruses (e.g. ZIKV, dengue, and influenza) cause large-scale alterations in transcriptional and metabolic expression to redistribute host-cell resources for replication [Citation36,Citation53]. Here, we found that ZIKV infection rewired the transcriptional structures of HBMECs but caused modest alterations in gene expression. In accordance with our findings, transcriptional alterations caused by ZIKV infection in myeloid dendritic cells, plasmacytoid dendritic cells, microglial (293FT) cells lines, and macrophage (THP-1) cell lines were also modest [Citation4,Citation36]. The activation of immune response is essential for host defense against viral infection, and clearance of existing infection. On the other hand, viruses have strategies to evade the host immune system. Indeed, ZIKV evades antiviral response through synergistic collaborative efforts among multiple nonstructural proteins (e.g. NS1, NS4B and NS5) in vitro and in vivo [Citation54,Citation55]. Here, we found that ZIKV evaded dysregulation of immune response genes and interacted with the upstream of genes in TVC7. Further experiments will be required to determine how these interactions contribute to the attenuated immune response upon ZIKV infection.
An appealing option for the development of antivirals is to target host proteins that are vital to viral infection. However, not all host proteins are druggable. By cross-referencing genes in TVC7 with DGIdb [Citation31], CYP1A2, E2F7, NRG1, PPP1R1B and TLR7 were identified as druggable targets for antivirals. TLR7, an intracellular sensor of single-stranded viral RNA (ssRNAs), plays an important role in mounting defense response against viral infections (e.g. Sendai virus, vesicular stomatitis virus, WNV, Enterovirus 71, influenza virus) [Citation56]. TLR7 agonist Imiquimod, also identified in this work, suppresses ZIKV replication in human fetal cardiac mesenchymal stromal cells in a dose-dependent manner [Citation40]. Known anti-ZIKV agents chloroquine and hydroxychloroquine repress ssRNAs-mediated activation of TLR7 signalling and the production of IFN-α [Citation57]. In this work, TLR7 loss of function weakly decreased ZIKV replication in HBMECs at 48 hpi compared with the control. Thus, the disruption of TLR7 signalling pathways might affect ZIKV replication. CYP1A2 inhibitor fluvoxamine/SAM001246977, estradiol-valerate/SAM002564206 (annotated in DrugBank) suppress ZIKV infection in human hepatoma cell Huh7 in a dose-dependent manner [Citation50]. Neuregulin 1 (NRG1) is one of four proteins in the neuregulin family (NRG1 through NRG4) that act on the EGFR family of receptors. HCV infection or the presence of the HCV sub-genomic replicon results in an enhanced expression of NRG1 at the transcript and protein level [Citation26]. Increased production of NRG1 results in an increased abundance of viral RNA, and neutralization of NRG1 substantially reduces the number of HCV-NS5A-expressing cells [Citation26]. Here, the knockdown of NRG1 expression reduced intracellular ZIKV RNA levels. Therefore, these druggable targets identified in this work could be used for computer-aided drug design or drug repositioning to discover novel anti-ZIKV agents.
Repositioning existing drugs is one of the most efficient ways to develop novel antivirals, as it is suitable for rapid translation to clinical use. Based on two distinct approaches, Signature-based drug repositioning and cross-reference with DGIdb, our work identified 74 approved drugs with the potential to inhibit ZIKV infection. The method based on drug-gene (protein or target) interaction databases found more known antivirals than signature-based drug repositioning. We are convinced that these agents could be repurposed for the administration of ZIKV infection, for 20 of the 74 drugs have been reported to inhibit ZIKV replication. Two drugs (Luminespib/NVP-AUY922, L-161982) with structures similar to known antivirals reduced ZIKV titres and viral replication in cellular assays. Luminespib/NVP-AUY922 is an HSP90 inhibitor. HSP90 antibodies block DENV infectivity in neuroblastoma cells [Citation58]. Here, Luminespib/NVP-AUY922 was identified as an antiviral based on signature-based drug repositioning and suppressed ZIKV infection in a cellular assay. Moreover, some genes in TVC7 are downstream of HSP90 in the reconstructed regulatory network ((d)). These suggest that inhibition of HSP90 by Luminespib/NVP-AUY922 may lead to the inhibition of gene expression in TVC7 and ZIKV infection. L-161982, a prostaglandin E2 (PGE2) receptor 4 (EP4) antagonist, induces apoptosis and inhibits prostaglandin E2-induced proliferation [Citation59]. PGE2 receptor EP2 or EP4 antagonist block dengue virus-induced dendritic cell migration and their combination have a synergistic effect [Citation60]. EP2 inhibitor PF-04418948 promotes AXL degradation [Citation44]. In summary, by exploring Drug-Targets/Signatures-ZIKV interactions, our work identified not only 22 known anti-ZIKV agents but also two approved drugs that could be repurposed as anti-ZIKV agents.
Despite the methods utilized in this work's effectiveness, several limitations should be addressed for the future. Firstly, studies related to the limited temporal resolution remain unknown about the structure and dynamics of host transcriptome during viral infection. High-resolution temporal transcriptome response to viral infection in host cells or tissues may obtain comprehensive and accurate insights. Single-cell RNAseq appears to be a more favourable option as it can dissect the viral infection process and host interactions at a higher resolution than that can be achieved with bulk RNAseq [Citation3]. It’s difficult to compare viral genome content between individual cells for that there will be cells at many stages of viral replicative cycle in theory. Conventional bulk RNA-seq captures measure gene expression during viral infection and provides statistical comparisons at the population level. Secondly, gene expression profiling usually displays significant differences between different cell types. The robustness of gene expression patterns during ZIKV infection should be verified on other cells. The discovery of potential antivirals depends on the capacity of drug signature databases (e.g. CMAP and LINCS) and drug-gene (protein or target) interaction databases (e.g. DrugBank and DGIdb).
In this work, we identified potential antivirals based on signature-based drug repositioning and cross-reference with drug-target databases. Candidate drugs with molecular structures similar to those of known anti-ZIKV drugs were evaluated for antiviral activity at the cellular level. However, we cannot ensure that all selected drugs are effective. The following are some possible reasons. (1) Due to the drugs’ pleiotropic nature, the intersection of their target range with key viral host factors may not be ideal; (2) In drug-target database, the activity of key host factors were indirectly affected by drugs; (3) Since only a limited number of genes were used as signatures in CMAP and LINCS in response to drug treatment, key host factors may not be genes that respond most strongly to drug treatment; (4) The cells tested in CMAP and LINCS were not the same as those (HBMECs) used in our experiments; (5) even if the drug candidates are structurally similar to known anti-ZIKV drugs, differences in physiochemical properties such as number of rotatable bonds, number of H-bond donor and acceptors present, partition coefficient, and blood–brain permeation may ultimately affect the antiviral activity of drugs.
Conclusions
Here, not only does our study shed new light on ZIKV infection mechanisms but also generated therapeutic opportunities for drug repurposing. Our data-driven approaches based on the temporal transcriptome have identified 5 druggable targets and 22 known anti-ZIKV agents, as well as 2 promising anti-ZIKV agents. Furthermore, our strategies should be useful for other pathogens causing pandemics and the rapid development of antivirals.
Acknowledgments
We thank Prof. Cui Min (State Key Laboratory of Agricultural Microbiology, College of Veterinary Medicine, Huazhong Agricultural University, Wuhan, China) for providing us with Human brain endothelial cells (HBMECs, passage 6, 10). N.Z. and Z.Z, conceived the project and designed the study. N.Z. wrote the paper. N.Z. designed and performed bioinformatics, drug repurposing, cheminformatics analysis and interpreted results. Z.T., N.Z., J.W., SZ., Y.L., J.M., C.J., designed and performed cell and viral experiments. N.Z. Z.Z., B.L. Z.T. and H.W. contributed to discussion and reviewed and edited the manuscript. All authors reviewed, edited the manuscript and approved the final manuscript. N.Z. and Z.Z. supervised the project.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Morrison TE, Diamond MS. Animal models of Zika virus infection, pathogenesis, and immunity. J Virol. 2017;91:e00009-17.
- Tan LY, Komarasamy TV, James W, et al. Host molecules regulating neural invasion of Zika virus and drug repurposing strategy. Front Microbiol. 2022;13:743147.
- Zanini F, Pu S-Y, Bekerman E, et al. Single-cell transcriptional dynamics of flavivirus infection. eLife. 2018;7:e32942.
- Sun X, Hua S, Chen H-R, et al. Transcriptional changes during naturally acquired Zika virus infection render dendritic cells highly conducive to viral replication. Cell Rep. 2017;21:3471–3482.
- Wouters OJ, McKee M, Luyten J. Estimated research and development investment needed to bring a new medicine to market, 2009-2018. JAMA. 2020;323:844.
- Low ZY, Farouk IA, Lal SK. Drug repositioning: New approaches and future prospects for life-debilitating diseases and the COVID-19 pandemic outbreak. Viruses-Basel. 2020;12:1058.
- Lamb J, Crawford ED, Peck D, et al. The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935.
- Subramanian A, Narayan R, Corsello SM, et al. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell. 2017;171:1437–1452.e17.
- Keenan AB, Jenkins SL, Jagodnik KM, et al. The library of integrated network-based cellular signatures NIH program: system-level cataloging of human cells response to perturbations. Cell Syst. 2018;6:13–24.
- Belyaeva A, Cammarata L, Radhakrishnan A, et al. Causal network models of SARS-CoV-2 expression and aging to identify candidates for drug repurposing. Nat Commun. 2021;12:1–13.
- Mladinich MC, Schwedes J, Mackow ER. Zika virus persistently infects and is basolaterally released from primary human brain microvascular endothelial cells. mBio. 2017;8:e00952-17.
- Lum F-M, Narang V, Hue S, et al. Immunological observations and transcriptomic analysis of trimester-specific full-term placentas from three Zika virus-infected women. Clin Transl Immunol. 2019;8:e01082.
- Ewels PA, Peltzer A, Fillinger S, et al. The nf-core framework for community-curated bioinformatics pipelines. Nat Biotechnol. 2020;38:276–278.
- Ernst J, Bar-Joseph Z. STEM: A tool for the analysis of short time series gene expression data. BMC Bioinformatics. 2006;7:191.
- Jung I, Jo K, Kang H, et al. Timesvector: a vectorized clustering approach to the analysis of time series transcriptome data from multiple phenotypes. Bioinformatics. 2017;33:3827–3835.
- Savidis G, McDougall WM, Meraner P, et al. Identification of Zika virus and dengue virus dependency factors using functional genomics. Cell Rep. 2016;16:232–246.
- Petrova E, Gracias S, Beauclair G, et al. Uncovering flavivirus host dependency factors through a genome-wide gain-of-function screen. Viruses. 2019;11:68.
- Lim Y-S, Ngo HTT, Lee J, et al. ADP-ribosylation factor-related protein 1 interacts with NS5A and regulates hepatitis C virus propagation. Sci Rep. 2016;6:31211.
- Leier HC, Weinstein JB, Kyle JE, et al. A global lipid map defines a network essential for Zika virus replication. Nat Commun. 2020;11:3652.
- Tsukada H, Ochi H, Maekawa T, et al. A polymorphism in MAPKAPK3 affects response to interferon therapy for chronic hepatitis C. Gastroenterology. 2009;136:1796–1805.e6.
- Aviner R, Frydman J. Proteostasis in viral infection: unfolding the complex virus–chaperone interplay. Cold Spring Harb Perspect Biol. 2020;12:a034090.
- Li J, Ding SC, Cho H, et al. A short hairpin RNA screen of interferon-stimulated genes identifies a novel negative regulator of the cellular antiviral response. mBio. 2013;4:e00385-13.
- Liao C, Zhou Q, Zhang Z, et al. Epstein-barr virus-encoded latent membrane protein 1 promotes extracellular vesicle secretion through syndecan-2 and synaptotagmin-like-4 in nasopharyngeal carcinoma cells. Cancer Sci. 2020;111:857–868.
- Luo W-W, Lian H, Zhong B, et al. Krüppel-like factor 4 negatively regulates cellular antiviral immune response. Cell Mol Immunol. 2016;13:65–72.
- Marcken M de, Dhaliwal K, Danielsen AC, Gautron AS, Dominguez-Villar M. TLR7 and TLR8 activate distinct pathways in monocytes during RNA virus infection. Sci Signal. 2019;12:eaaw1347.
- Stindt S, Cebula P, Albrecht U, et al. Hepatitis C virus activates a neuregulin-driven circuit to modify surface expression of growth factor receptors of the ErbB family. PLoS ONE. 2016;11:e0148711.
- Yang X, Lian X, Fu C, et al. HVIDB: A comprehensive database for human–virus protein–protein interactions. Brief Bioinform. 2021;22:832–844.
- Teng Y, Liu S, Guo X, et al. An integrative analysis reveals a central role of p53 activation via MDM2 in Zika virus infection induced cell death. Front Cell Infect Microbiol. 2017;7:327.
- Yang W, Wu Y-H, Liu S-Q, et al. S100a4+ macrophages facilitate Zika virus invasion and persistence in the seminiferous tubules via interferon-gamma mediation. PLOS Pathog. 2020;16:e1009019.
- Liu S, DeLalio LJ, Isakson BE, et al. AXL-mediated productive infection of human endothelial cells by Zika virus. Circ Res. 2016;119:1183–1189.
- Freshour SL, Kiwala S, Cotto KC, et al. Integration of the drug–gene interaction database (DGIdb 4.0) with open crowdsource efforts. Nucleic Acids Res. 2021;49:D1144–D1151.
- Oo A, Teoh BT, Sam SS, et al. Baicalein and baicalin as Zika virus inhibitors. Arch Virol. 2019;164:585–593.
- Zhang S, Yi C, Li C, et al. Chloroquine inhibits endosomal viral RNA release and autophagy-dependent viral replication and effectively prevents maternal to fetal transmission of Zika virus. Antiviral Res. 2019;169:104547.
- Persaud M, Martinez-Lopez A, Buffone C, et al. Infection by Zika viruses requires the transmembrane protein AXL, endocytosis and low pH. Virology. 2018;518:301–312.
- Barrows NJ, Campos RK, Powell ST, et al. A screen of FDA-approved drugs for inhibitors of Zika virus infection. Cell Host Microbe. 2016;20:259–270.
- Tiwari SK, Dang J, Qin Y, et al. Zika virus infection reprograms global transcription of host cells to allow sustained infection. Emerg Microbes Infect. 2017;6:e24.
- Kuivanen S, Bespalov MM, Nandania J, et al. Obatoclax, saliphenylhalamide and gemcitabine inhibit Zika virus infection in vitro and differentially affect cellular signaling, transcription and metabolism. Antiviral Res. 2017;139:117–128.
- Kumar A, Liang B, Aarthy M, et al. Hydroxychloroquine inhibits Zika virus NS2B-NS3 protease. ACS Omega. 2018;3:18132–18141.
- Duarte G, Moron AF, Timerman A, et al. Zika virus infection in pregnant women and microcephaly. Rev Bras Ginecol E Obstetrícia. 2017;39:235–248.
- Rossi F, Josey B, Sayitoglu EC, et al. Characterization of Zika virus infection of human fetal cardiac mesenchymal stromal cells. PLoS One. 2020;15:e0239238.
- Li Z, Yao F, Xue G, et al. Antiviral effects of simeprevir on multiple viruses. Antiviral Res. 2019;172:104607.
- Chuang F-K, Liao C-L, Hu M-K, et al. Antiviral activity of compound l3 against dengue and Zika viruses in vitro and in vivo. Int J Mol Sci. 2020;21:4050.
- Grady S, Pinto A, Hassert M, et al. Tamoxifen as a Zika virus therapeutic. FASEB J. 2021;35:S1.
- Pan T, Peng Z, Tan L, et al. Nonsteroidal anti-inflammatory drugs potently inhibit the replication of Zika viruses by inducing the degradation of AXL. J Virol. 2018;92:e01018-18.
- Richter M, Boldescu V, Graf D, et al. Synthesis, biological evaluation, and molecular docking of combretastatin and colchicine derivatives and their hCE1-activated prodrugs as antiviral agents. ChemMedChem. 2019;14:469–483.
- Li M, Zhang D, Li C, et al. Characterization of Zika virus endocytic pathways in human glioblastoma cells. Front Microbiol. 2020;11:242.
- Medigeshi GR, Kumar R, Dhamija E, et al. N-desmethylclozapine, fluoxetine, and salmeterol inhibit postentry stages of the dengue virus life cycle. Antimicrob Agents Chemother. 2016;60:6709–6718.
- Guo J, Jia X, Liu Y, et al. Inhibition of na+/K+ ATPase blocks Zika virus infection in mice. Commun Biol. 2020;3:1–8.
- Han Y, Mesplède T, Xu H, et al. The antimalarial drug amodiaquine possesses anti-ZIKA virus activities. J Med Virol. 2018;90:796–802.
- Pascoalino BS, Courtemanche G, Cordeiro MT, et al. Zika antiviral chemotherapy: identification of drugs and promising starting points for drug discovery from an FDA-approved library. F1000Res. 2016;5:2523.
- Mounce BC, Cesaro T, Carrau L, et al. Curcumin inhibits Zika and chikungunya virus infection by inhibiting cell binding. Antiviral Res. 2017;142:148–157.
- Li Z, Lang Y, Sakamuru S, et al. Methylene blue is a potent and broad-spectrum inhibitor against Zika virus in vitro and in vivo. Emerg Microbes Infect. 2020;9:2404–2416.
- Thaker SK, Chapa TJ, Garcia G, et al. Differential metabolic reprogramming by Zika virus promotes cell death in human versus mosquito cells. Cell Metab. 2019;29:1206–1216.
- Kwock JT, Handfield C, Suwanpradid J, et al. IL-27 signaling activates skin cells to induce innate antiviral proteins and protects against Zika virus infection. Sci Adv. 2020;6:eaay3245.
- Wu Y, Liu Q, Zhou J, et al. Zika virus evades interferon-mediated antiviral response through the co-operation of multiple nonstructural proteins in vitro. Cell Discov. 2017;3:17006.
- Sartorius R, Trovato M, Manco R, et al. Exploiting viral sensing mediated by toll-like receptors to design innovative vaccines. Npj Vaccines. 2021;6:1–15.
- Gies V, Bekaddour N, Dieudonné Y, et al. Beyond anti-viral effects of chloroquine/hydroxychloroquine. Front Immunol. 2020;11:1409.
- Lubkowska A, Pluta W, Strońska A, et al. Role of heat shock proteins (HSP70 and HSP90) in viral infection. Int J Mol Sci. 2021;22:9366.
- Li X, Yang B, Han G, et al. The EP4 antagonist, L-161,982, induces apoptosis, cell cycle arrest, and inhibits prostaglandin E2-induced proliferation in oral squamous carcinoma Tca8113 cells. J Oral Pathol Med. 2017;46:991–997.
- Wu W-L, Ho L-J, Chang D-M, et al. Triggering of DC migration by dengue virus stimulation of COX-2-dependent signaling cascades in vitro highlights the significance of these cascades beyond inflammation. Eur J Immunol. 2009;39:3413–3422.

