ABSTRACT
Background & Aims
The WHO declared to eliminate hepatitis B virus (HBV) by 2030. However, an increasing number of patients are presenting with low-level viremia (LLV) with the widespread use of antiviral medications. The diagnostic efficiency and coverage area of HBV infection are low. Hence, this study intended to drive the HBV infection detection to effectively adaptable for any small to medium-sized laboratory or field survey.
Methods
We established, optimized, and evaluated a colloidal gold test strip for detection of HBV DNA based on CRISPR/Cas13a combined with recombinase-aided amplification (RAA) technology. Furthermore, 180 HBV-infected patients (including patients with different viral loads, LLV patients and dynamic plasma samples of patients on antiviral therapy) were enrolled for clinical validation.
Results
The strip detection of HBV DNA was established based on RAA-CRISPR-Cas13a technology with a sensitivity of 101 copies/μL and a specificity of 100%. HBV DNA gradient concentration plasmids and clinical samples were effectively identified by this approach. The positive coincidence rate for LLV patients was 87%, while the negative coincidence rate was 100%. The positive coincidence rate reached 100% in LLV patients (viral loading >100 IU/mL). The sensitivity, specificity, positive predictive agreement (PPA) and negative predictive agreement (NPA) values of dynamic plasma detection in patients on antiviral therapy were 100%, 92.15%, 93.75%, and 100%, respectively.
Conclusions
We develop rapid and portable RAA-CRISPR/Cas13a-based strip of HBV DNA detection for LLV patients. This study provides a visual and faster alternative to current PCR-based diagnosis for HBV infection.
1. Introduction
Hepatitis B virus (HBV) infection is a severe public health problem. It is estimated that there are 257–292 million people with HBV infections worldwide; more than 95% of these patients live in low- and middle-income countries, and only 12–25% of these patients are eligible for diagnosis and anti-HBV treatment [Citation1, Citation2]. Furthermore, after long-term standardized treatment with first-line antiviral drugs, persistent or intermittent low levels of positive HBV DNA in plasma, known as low-level viremia (LLV), have been detected in an increasing number of patients [Citation3]. This is becoming a key part of and new challenge in the treatment and management of chronic hepatitis B (CHB). Therefore, the World Health Organization (WHO) has developed a strategy to eliminate HBV as a public health threat by 2030, and one of the main objectives is to improve the efficiency and coverage of HBV testing [Citation4].
As a direct indicator of viral replication and infectivity, HBV DNA detection plays a crucial role in judging viral infectivity and antiviral effects, especially in evaluating the endpoint of antiviral therapy in LLV patients. Although real-time polymerase chain reaction (RT–qPCR)-based detection methods have been widely used for HBV DNA detection with superior performance [Citation5], they have shortcomings. These limitations are mainly associated with the much longer turnaround time for the results, the requirement for large-scale equipment and a shortage of professional and technical personnel [Citation6, Citation7]. Furthermore, studies have shown that low awareness of the disease and the high cost of testing contribute to the lack of HBV testing in low- and middle-income countries [Citation8, Citation9]. Therefore, the detection of HBV DNA needs to be further improved in terms of detection time and portability.
The test strip, as a detection method that is carried out at a sampling site and can quickly obtain detection results based on monoclonal antibody technology, colloidal gold labeling technology and immunochromatography technology, has received widespread attention. Compared with traditional detection methods, the test strip has the advantages of simple operation, fast response, and portable equipment, so it has been rapidly developed and widely used in disease diagnosis [Citation10]. However, the test strip for HBV mostly depends on serological markers such as HBsAg [Citation11, Citation12], while it for HBV DNA is vacant. Notably, some emerging nucleic acid molecular diagnostics have met requirements for sensitivity, specificity, and low cost for the test strip applications and thus have become a trend in point-of-care testing (POCT) development.
In the past decade, clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated systems (Cas), a new nucleic acid detection technology, have attracted considerable attention and made great progress due to their unique properties of high sensitivity and high specificity. A typical example was reported by Feng Zhang et al. who used CRISPR/Cas technology combined with recombinase polymerase amplification (RPA) for nucleic acid detection in a practical molecular diagnostic technology platform called SHERLOCK [Citation13]. Subsequently, RPA-CRISPR/Cas technology has been widely used to detect severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), Zika virus, and Dengue virus [Citation14, Citation15]. Furthermore, combined with a lateral flow strip, the CRISPR system can take advantage of the rapid and portable features of POCT to achieve high-sensitivity and high-specificity detection of pathogenic nucleic acids without professional personnel; this system has also been used to successfully detect SARS-CoV-2 [Citation16, Citation17]. Thus, the combination of a lateral flow strip and CRISPR/Cas technology might provide a new platform for the rapid detection of HBV DNA.
To simplify the operation, shorten the time, and reduce the cost, this study intended to develop a rapid and portable HBV DNA detection test strip based on recombinase-aided amplification (RAA)-CRISPR/Cas13a, which can achieve high-sensitivity and high-specificity detection in LLV patients and can assist rapid and portable HBV DNA detection of clinical use and in low-income countries.
2. Materials and methods
2.1 Ethics statement
The current investigation's procedure complied with the 1975 Declaration of Helsinki ethical criteria and the 1983 amendments, as evidenced by a priori clearance by Beijing Youan Hospital's human research committee (Ethical code: LL-2020-167-K). All the patients gave informed consent.
2.2 Clinical samples
At Beijing Youan Hospital, Capital Medical University, 180 HBV-infected patients and 25 healthy individuals were enrolled (their clinical characteristics are shown in Supplementary Table 1), and their plasma samples were measured using HBV DNA ultrasensitive qPCR (Abbott M2000 RealTime PCR System, USA). Fifteen patients (no HBV DNA was detected), 42 patients with different viral loads, 100 patients with LLV, and 23 patients on antiviral treatment comprised the 180 patients. In addition, 111 dynamic plasma samples from the treatment course were collected from 23 individuals on antiviral medication. The CHB patients and LLV patients included in the study met the diagnostic criteria of the American Association for the Study of Liver Diseases (AASLD) guidelines (LLV is defined as HBV DNA persistently or intermittently greater than the lower limit of detection but less than 2,000 IU/mL) [Citation18]. Ten patients with HCV-, HEV- and HIV-positive nucleic acids were selected for method specificity evaluation. The Laboratory Department of Beijing Youan Hospital tested for HBV DNA and associated blood indicators in all the collected samples. Patient information was obtained from their medical records. Each patient's plasma was collected and kept at −80°C until sample processing. All the samples were anonymized before being tested in the laboratory.
2.3 Synthesis of plasmids, primers, and crRNAs
The HBV plasmid used in this work, along with the crRNA in the Cas13a-crRNA assay system, can cover eight different HBV genotypes. We downloaded approximately 10000 HBV DNA sequences that had been updated at the time from the HBV database [Citation19] (https://hbvdb.lyon.inserm.fr/HBVdb/HBVdbIndex). Approximately 10000 HBV P-region sequences were compared using Jalview software, and the results are displayed in Supplementary Figure 1. Following the comparison, an HBV DNA sequence of 134 bp with more than 95% conservation was chosen (Supplementary Figure 1), followed by the construction of the HBV-positive control plasmid and the creation of the appropriate crRNA and RAA primers based on the conserved sequences (Supplementary Tables 2 and 3). The crRNA and RAA primers were manufactured by Sangon Biotech (Shanghai) Co., Ltd. A plasmid containing the partial HBV genome (GenBank ID: LC456126.1 site: 625-758) and pUC57 served as the templates to construct an artificial HBV DNA standard (Bomide Biotechnology Co., Ltd).
2.4 Plasma HBV DNA extraction
HBV DNA was extracted from plasma samples using the TIANamp Virus DNA/RNA Kit (Tiangen Biotech (Beijing) Co., Ltd.). The extraction procedure was followed exactly as directed by the kit. The extraction process was as follows: 20 μL Proteinase K, 200 μL plasma and 200 μL carrier RNA working solution were added to a 1.5 mL centrifuge tube, in that order, and incubated at 56°C for 15 min. Next, 250 μL anhydrous ethanol was added to the centrifuge tube; the solution was transferred to a CR2 adsorption column, and the waste liquid was centrifuged. A buffer solution, rinse solution (once), and anhydrous ethanol were then added and centrifuged for 1 min at 8000 rpm, and the waste liquid was removed. The adsorption film was fully dried by centrifugation at 12000rpm for 3 min. The adsorption column was placed in an RNase-free centrifuge tube, and 20 μL RNase-free ddH2O was added to the centre of the adsorption membrane, which was then left at room temperature for 5 min before being centrifuged at 12,000 rpm for 1 min to obtain a 20 μL HBV DNA extract. We performed quality control experiments on HBV DNA extracts with the aim of demonstrating that the background or nonspecific substrates of the reaction had no effect on the assay. (Supplementary Figure 2).
2.5 Preparation and purification of Cas13a-crRNAs
Complete crRNA sequences were generated as single-stranded DNA and then PCR-amplified with an added T7 promoter sequence to produce crRNAs. Using the HiScribe T7 Quick High Yield RNA Synthesis Kit (New England Biolabs, Inc.), two complementary pairs of crRNA DNA primers (final concentrations 10 nM) were synthesized and treated overnight with T7 polymerase at 37°C (New England Biolabs, Inc.). An RNA Rapid Concentration Purification Kit was then used to purify the crRNAs (Sangon Biotech, Inc.).
2.6 Recombinase-aided amplification (RAA)
Isothermal amplification of the plasmid or extraction of HBV DNA was performed according to the instructions of the RAA kit (Hangzhou Zhongshan Bio-Sci&Tech Co., Ltd.). According to the reaction system, 13.5 μL of water, 25 μL of Buffer A, and 2 μL of upstream and downstream primers (10 nM) were combined uniformLy into the detecting unit tube containing dry reaction powder. Then, a 5 μL DNA sample was placed into the detection unit tube; 2.5 μL Buffer B was then added to the detection unit's tube cover, the tube cover was closed, and the mixture was completely mixed 5–6 times with gentle shaking upside down, centrifuged at low speed for 10 sec, and put in a metal bath at 39°C for 30 min to acquire the amplified products.
2.7 Dna agarose gel electrophoresis
To create a 1% agarose gel, 1 g agarose (Solarbio®) was combined with 100 mL of 1×TAE solution (Bomide Biotechnology Co., Ltd.) and heated until dissolved. The mixture was chilled to 50-60°C before adding 10 μL GoldenViewTM nucleic acid dye (Bomide Biotechnology Co., Ltd). The gel was placed in an electrophoresis tank containing 1×TAE Buffer, 5 μL PCR amplification product, and 1 μL 6× RNA/DNA loading buffer (Bomide Biotechnology Co., Ltd), and the gel hole was filled with the mixture. The first hole was filled with 5 μL of the BM5000 DNA Marker (Bomide Biotechnology Co., Ltd.). After electrophoresis at 170 V for approximately 20 min, the gel blocks were removed and placed into the gel imager (Bio–Rad, Inc.) for examination and image preservation.
2.8 Cas13a-crRNA nucleic acid fluorescence detection
The following procedure was followed for detection. In nuclease assay buffer, 1 μL of RNase inhibitor (New England Biolabs, Inc.), 1 μL LwCas13a, 1.5 μL crRNA, 2 μL NTP Mix (New England Biolabs, Inc.), 0.5 μL of T7 RNA Polymerase (New England Biolabs, Inc.), 0.5 μL of HEPES buffer solution (ThermoFisher Scientific, Inc., USA), 0.25 μL MgCl2 solution (1 M), 2.5 μL quenched fluorescence RNA reporter (RNAse Alert, Thermo Scientific, Waltham, MA, USA), 5 μL of target nucleic acid, and 10.75 μL of RNase-free water were added. The reactions were carried out at 37°C for 1 h, and the fluorescence signal was collected every 2 min on an ABI 7500 Fast DX (Applied Biosystems, Foster City, CA, USA).
2.9 Cas13a-crRNA nucleic acid strip detection
The test (T) and control (C) lines were formed by spraying 1 mg/mL streptavidin (Thermo Fisher Scientific, Waltham, MA, USA) and 1.5 mg/mL goat antirabbit immunoglobulin G (IgG) (Solarbio, Beijing, China) onto nitrocellulose membranes. A colloidal gold-labeled rabbit anti-FAM antibody (Sangon Biotech, Shanghai, China) was sprayed onto a conjugate pad formed from a piece of glass microfiber filter at a final concentration of 0.01 mg/mL.
The following procedure was followed for detection: In nuclease assay buffer, 2 μL of RNase inhibitor (New England Biolabs, Inc.), 2 μL LwCas13a, 3 μL crRNA, 4 μL NTP Mix (New England Biolabs, Inc.), 1 μL of T7 RNA Polymerase (New England Biolabs, Inc.), 1 μL of HEPES buffer solution (ThermoFisher Scientific, Inc., USA), 0.5 μL of MgCl2 solution (1 M), 2 nM reporter RNA (5’/6-FAM/UUUUUUUUUUUUUUUUUUUU-Bio/3’), 5 μL of target nucleic acid, and 26.5 μL of RNase-free water were added. The reactions were carried out at 37°C for 30 min.
The reaction complex was added to the strip, and the results were evident after 3 min. There were strips on both the control line (C) and test line (T), which were negative results. A single band on the control line (C) indicated a positive result. The ImageJ analysis tool was used to examine the band strength.
2.10 Assessment of detection capabilities
Evaluation of specificity: After constructing different viral plasmids (HCV, HDV, HEV, HIV), RAA was performed using 103 copies/μL plasmid as a template, and the products were subjected to a Cas13a-crRNA detection reaction. The products of HCV, HEV, HIV plasma RNA extraction, reverse transcription, and RAA were used to further evaluate the specificity of the method. Evaluation of sensitivity: HBV plasmids diluted in gradient concentrations (104, 103, 102, 101, 100 copies/μL) were amplified by RAA for Cas13a-crRNA detection. Clinical samples with different concentrations (104, 103, 102, 101, < 10 IU/mL) were further evaluated for limits of detection (LoDs) after HBV DNA extraction and RAA. Evaluation of repeatability: An HBV plasmid of 103 copies/μL was amplified by RAA and then repeatedly detected 20 times, and its endpoint fluorescence value was analyzed by using SIMCA.
2.11 Optimization of reaction conditions
The Cas13a-crRNA-fluorescence assay signal was utilized to optimize the duration and temperature throughout the process, and the endpoint fluorescence value was chosen for comparison. The samples of optimization process are all 105 IU/mL blood samples. The optimization process controlled a single variable, including time (5, 15, 30, 60 min) and temperature (25, 37, 56, 95°C) of sample lysis, sample storage days (0, 1, 3, 5, 7 days), RAA reaction time (5, 15, 30, 45, 60 min), temperature (25, 33, 36, 39, 42, 45°C) and template volume (2, 4, 6, 10 μL), CRISPR reaction time (10, 20, 30 min) and temperature (25, 28, 31, 34, 37, 40, 43°C), and comparison of plasma and serum.
2.12 Statistical analysis
The Cas13a-crRNA-fluorescence assay signal was expressed as the mean of ≥3 independent reactions ± SD. GraphPad Prism software version 8.0 (GraphPad, Inc., La Jolla, CA, USA) was used to determine the means and SDs to calculate the analysis of variance (ANOVA). The paired t test was used to detect mean differences in quantification. All statistical tests were two-sided, and statistical significance was defined as P < 0.05.
3. Results
3.1 Construction of the Cas13a-crRNA detection system for HBV DNA
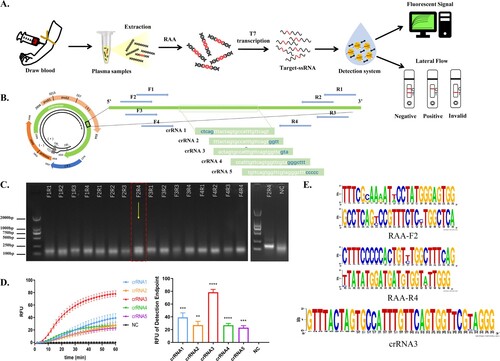
The process of HBV DNA detection via CRISPR/Cas13a-crRNA involved collecting plasma from HBV patients and extracting HBV DNA following sample processing. After RAA, target single-stranded RNA (ssRNA) was transcribed by using T7 transcriptase. When crRNA binds to ssRNA, the Cas13a accidental cleavage activity is activated, cleaving the reporter RNA, which may be detected using a fluorescence signal or a test strip (). After sequence comparison, the conserved target HBV sequence (approximately 200 bp) was chosen, and 5 crRNAs and 3 pairs of RAA primers were built based on this sequence (), allowing Cas13a-crRNA to detect numerous HBV subtypes. The agarose gel electrophoresis assay results revealed that primer F2R4 had the greatest amplification efficiency and was employed in further studies (). The findings of Cas13a-crRNA fluorescence detection revealed that crRNA3 was the most effective, with the highest endpoint fluorescence value and the best fluorescence efficiency (). This result was validated by conservation analysis of the screened RAA primers and crRNA (). Hence, we were able to effectively establish a Cas13a-crRNA detection system for HBV DNA.
Figure 1. Construction of a Cas13a-crRNA Detection System for HBV DNA. Data are representative of at least three independent experiments. (****<0.0001, ***<0.001, **<0.01) (A) Process of the Cas13a-crRNA detection system for HBV DNA detection. (B) The schematic diagram of the design includes five crRNAs and four pairs of RAA primers, and the band (5' to 3') indicates the conserved region of the HBV sequence that was screened. (C) Screening of RAA primers. HBV-positive plasmids (103 copies/μL) were amplified by RAA, and the amplified products were subjected to DNA agarose gel electrophoresis. Then, the F2R4 primers screened out were further verified by using an HBV-positive plasmid (105 copies/μL). (D) Screening of crRNA. The two figures are the comparison of the dynamic fluorescence curve and end-point fluorescence value of different crRNAs. The chosen RAA primers amplified the HBV-positive plasmid (103 copies/μL) and served as a template for Cas13a-crRNA fluorescence detection to verify five crRNAs. (E) Conservation analysis of selected crRNA and RAA primers by WebLogo.
3.2 Capability of the Cas13a-crRNA system for detecting HBV DNA
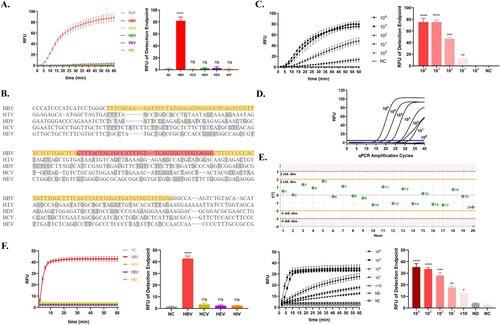
The capability of the Cas13a-crRNA detection system was assessed by determining the LoD, specificity, and repeatability. We compared the sequences of conserved regions of HBV with other common clinical viruses, such as hepatitis viruses, and constructed corresponding plasmids for Cas13a-crRNA detection. The findings revealed that HBV had the greatest terminal fluorescence detection value and the best fluorescence efficiency, with a detection specificity of 100% (). Moreover, the serially diluted HBV plasmids were amplified by RAA for Cas13a-crRNA detection. The Cas13a-crRNA detection system found a low LoD of 101 copies/μL (). When compared to qPCR, the standard approach for detecting HBV DNA in clinical practice, qPCR detection had the same degree of LoD but reacted more slowly (). For example, the Cas13a-crRNA method detected the fluorescence signal within 10 min of reaction, whereas qPCR could not. In addition, after 20 repeated trials, the Cas13a-crRNA detection system's standard deviation was lower, indicating superior repeatability (). Moreover, the constructed Cas13a-crRNA detection system can satisfy for the detection needs of clinical samples in terms of both specificity and sensitivity ().
Figure 2. Capability of the Cas13a-crRNA Detection System for HBV DNA. Data are representative of at least three independent experiments. (NC: negative control, ND: no detection for HBV DNA in HBV-infected patients, ****<0.0001, ***<0.001, **<0.01, *<0.05). (A) The specificity assessment of the Cas13a-crRNA detection system using HBV, HCV, HDV, HEV, and HIV plasmid (103 copies/μL). (B) The alignment of conserved regions of HBV with other common clinical viruses (HIV, HCV, HDV, and HEV). RAA primer and crRNA-binding regions are highlighted in yellow and red, respectively. (C) The sensitivity assessment of the Cas13a-crRNA detection system by HBV plasmids diluted in gradient concentrations (100∼104 copies/μL). (D) qPCR amplification curves of HBV plasmids at different concentrations (100∼106 copies/μL). (E) The repeatability assessment of the Cas13a-crRNA detection system by HBV plasmid (103 copies/μL) (Standard Deviation, Std. dev.). (F) The specificity assessment of the Cas13a-crRNA detection system by clinical samples (103 IU/mL) and the sensitivity assessment of the Cas13a-crRNA detection system by clinical samples with different concentrations (104, 103, 102, 101, <10 IU/mL).
3.3 Optimization of the Cas13a-crRNA detection system for HBV DNA
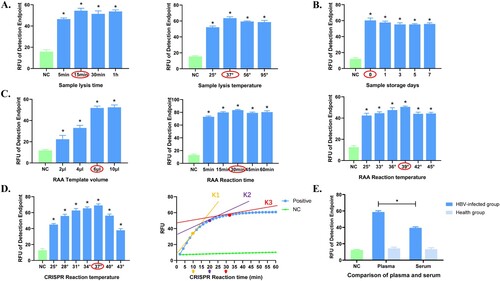
We optimized and compared the Cas13a-crRNA detection process by the fluorescence signal intensity at the end point as a scale. The best reaction for the sample lysis process was lysis at 37° for 15 min (), with the use of fresh plasma samples (). The optimal choice for RAA was 39° amplification for 30 min, using 6 μL of amplification template for maximal fluorescence efficiency (). The CRISPR reaction performed best at 37°, and the entire reaction process was finished in less than 30 min. At this time, the slope of the curve tangent was near 1 (). We also evaluated how the difference in plasma and serum affected the detection of HBV-infected individuals. The results showed that the effect of plasma was better (). Therefore, the Cas13a-crRNA detection system for HBV DNA was optimized and used in the subsequent step.
Figure 3. Optimization of the Cas13a-crRNA Detection System for HBV DNA. Data are representative of at least three independent experiments. (NC: negative control, *<0.05). (A) Optimization of sample lysis time (5, 15, 30, and 60 min) and sample lysis temperature (25, 37, 56, and 95°C). (B) Comparison of sample storage days (0, 1, 3, 5, and 7 days). (C) Optimization of RAA reaction time (5, 15, 30, 45, 60 min), temperature (25, 33, 36, 39, 42, 45°C) and template volume (2, 4, 6, 10 μL). (D) Optimization of CRISPR reaction time (10, 20, and 30 min) and temperature (25, 28, 31, 34, 37, 40, and 43°C). K1, K2 and K3 represent the slope of fluorescence curve at the time points of 10, 20 and 30 minutes of CRISPR reaction, reflecting the amplification efficiency. (E) Comparison of plasma and serum.
3.4 Cas13a-crRNA-Based strip assay for HBV DNA
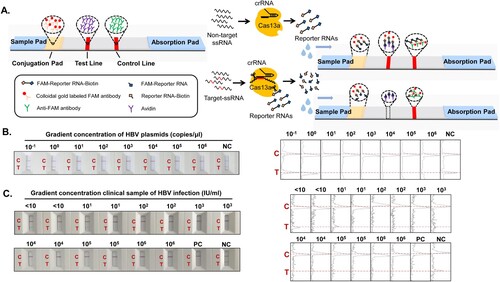
We used the Cas13a-crRNA system in conjunction with colloidal gold test strips to detect HBV DNA rapidly and portably. The following is the “elimination method” strip detection of HBV DNA principle (): The test strip was constructed containing a sample pad, conjugation pad (with colloidal gold-labeled FAM antibody), test line (with avidin), control line (with anti-FAM antibody), and absorption pad. The liquid dropped onto the test strip runs through the conjugation pad, detection line, and control line in that order, according to the test strip's siphon effect. When the Cas13a-crRNA complex detects the target nucleotide, Cas13a engages in trans-cleavage activity to cleave the reporter RNA (FAM-Reporter RNA-Biotin). The colloidal gold molecules are unable to remain on the test line due to cleavage of the reporter RNA, so no bands emerge (positive). Conversely, colloidal gold molecules reside on the test line due to the formation of “colloidal gold-labeled FAM antibody-FAM-Reporter RNA-Biotin–Avidin” complexes and thus appear as bands (Negative). Notably, the control line was coated with anti-FAM antibody and could bind to colloidal gold-labeled FAM antibody to display bands in either circumstance. The test strip was judged as invalid if no bands appeared. Subsequently, the method was validated using gradient concentrations of HBV plasmids () and clinical plasma samples with different viral loads (), all of which were positive.
Figure 4. Cas13a-crRNA-Based Strip Assay for HBV DNA. Data are representative of at least three independent experiments. (NC: negative control, PC: positive control). (A) Schematic diagram of the Cas13a-crRNA nucleic acid detection strip. (B) The gradient concentration of HBV plasmids (10-1∼106 copies/μL) was assayed with a Cas13a-crRNA nucleic acid detection strip. The signal intensities of the T and C bands were generated using ImageJ software. (C) Clinical samples of HBV infection with different viral loads (<10∼106 IU/mL, n=42) were assayed with a Cas13a-crRNA nucleic acid detection strip. The signal intensities of the T and C bands were generated using ImageJ software.
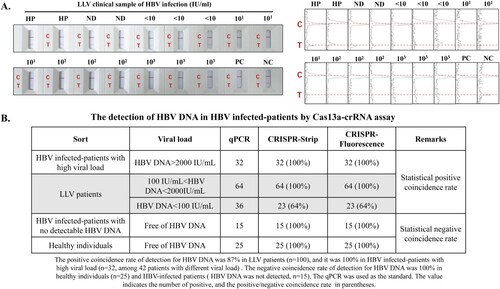
3.5 Cas13a-crRNA-assisted HBV DNA strip assay in LLV patients
To evaluate the validity of Cas13a-crRNA for strip detection, HBV-infected LLV patients were selected for clinical validation. The results indicated that healthy people and no-detection individuals had negative results, whereas LLV patients had positive results (). Moreover, according to a comparison of the three methods of qPCR, Cas13a-crRNA strip detection and Cas13a-crRNA fluorescence detection, the positive coincidence rate of Cas13a-crRNA strip detection in the detection of HBV DNA in LLV patients was approximately 87%, and the negative coincidence rate was 100% (). Notably, when HBV DNA levels in LLV patients exceeded 100 IU/mL, the positive coincidence rate of Cas13a-crRNA strip detection reached 100% ().
Figure 5. Figure 5. Cas13a-crRNA-Assisted HBV DNA Strip Assay in LLV Patients. Data are representative of at least three independent experiments. (NC: Negative Control, PC: Positive Control, HP: Healthy People, ND: No detection for HBV DNA in HBV-infected patients, <10: The content of HBV DNA in the sample was 10 IU/mL, 101: The content of HBV DNA in the sample was 101 IU/mL, 102: The content of HBV DNA in the sample was 102 IU/mL, 103: The content of HBV DNA in the sample was 103 IU/mL.). (A) LLV clinical samples (<10∼103 IU/mL) of HBV infection were assayed with a Cas13a-crRNA nucleic acid detection strip. The signal intensities of the T- and C-bands were generated using ImageJ software. (B) Analysis of positive/negative coincidence rate about Cas13a-crRNA assay for detection of HBV DNA in HBV-infected patients.
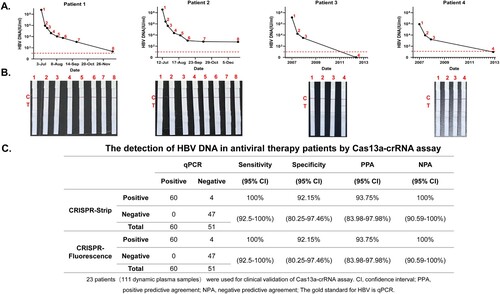
3.6 Cas13a-crRNA-assisted HBV DNA strip assay in antiviral therapy patients
To further evaluate the validity of Cas13a-crRNA for strip detection, the dynamic plasma samples of 23 CHB antiviral patients (111 samples) were chosen to increase the number of clinical samples for validation. The qPCR detection results showed the change trend in plasma HBV DNA at different time points in different patients taking antiviral drugs ( and Supplementary Figure 3). The Cas13a-crRNA strip detection results of each patient's corresponding dynamic time point were consistent with the qPCR results (). In addition, using qPCR as the standard for HBV DNA detection, the sensitivity, specificity, positive predictive agreement (PPA), and negative predictive agreement (NPA) values of Cas13a-crRNA strip detection for HBV DNA in the dynamic samples from CHB antiviral patients were 100%, 92.15%, 93.75% and 100%, respectively ().
Figure 6. Cas13a-crRNA-Assisted HBV DNA Strip Assay in Antiviral Therapy Patients. Data are representative of at least three independent experiments. (A) The dynamic line chart by GraphPad Prism shows the changes in HBV DNA levels (IU/mL) in patients treated with antiviral therapy. (B) Antiviral therapy dynamic plasma samples (IU/mL) were assayed with a Cas13a-crRNA nucleic acid detection strip. (C) Analysis of sensitivity, specificity, PPA, and NPA about Cas13a-crRNA assay for detection of HBV DNA in antiviral therapy patients.
4. Discussion
The new method for HBV DNA detection established and explored in this study reveals numerous distinct benefits for meeting the demands of detecting HBV-related diseases: (1) Rapid and portable HBV DNA detection was possible based on the combination of nucleic acid detection with the immunocolloidal gold chromatography technique. (2) HBV DNA detection based on the CRISPR/Cas13a system exhibits excellent specificity and sensitivity, and it can cover 8 genotypes of HBV (3) This method has great stability and predictive value for detecting HBV DNA in patients with LLV and in those receiving antiviral treatment. (4) The benefits of low cost and easy promotion can significantly alleviate the difficulties of uneven HBV diagnosis, low detection efficiency, and limited coverage.
The test strip application offers the most benefit in terms of rapidity and portability in the diagnosis and screening of HBV infection. Currently, in many poor countries and locations with high HBV incidence, qPCR and ELISA detection technologies may not be the ideal choices for HBV detection [Citation20, Citation21], resulting in low HBV detection efficiency and coverage due to space and time constraints. However, this issue could be overcome by the test strip, which is associated with fast results and portability and can be performed by nonlaboratory or nonmedical individuals and thus could be used for patient self-monitoring, and the results are accessible in a timely manner for evaluating illness and are particularly useful in remote rural locations [Citation22]. Hence, for this study, we picked the portable colloidal gold test strip, which is most widely used in POCT. Moreover, it considerably reduces the turnaround time for clinical samples, and test findings are easily available at the point of care, allowing for more timely clinical decision-making and treatment [Citation23]. We discovered that the approach established in this paper is faster than qPCR detection. Notably, most of the time is consumed by sample processing and this processing uses a little equipment. Fortunately, extracting HBV DNA in under 30 s is no longer a pipe dream [Citation24], and we may be able to shorten and optimize our procedure even more. We also discovered that plasma was more suitable than serum for the detection of HBV DNA in our study. We hypothesized that a small number of HBV particles in the blood may be wrapped together to form clots due to fibrin contraction and the aggregation of red blood cells during the coagulation process, resulting in a slightly lower HBV level in serum than in plasma. Moreover, other research has indicated that in clinical settings, plasma samples are better suited for liquid biopsy [Citation25]. In addition to the inexpensive cost of this test, the expense of acquiring and maintaining equipment, staff training, and needless patient transportation are significantly reduced [Citation22].
We applied CRISPR/Cas13a technology to detect HBV DNA to achieve high sensitivity and high specificity. CRISPR is an acquired immune system in most bacteria and archaea that protects them from viral invasion. It can recognize and incorporate foreign viral gene segments and then be transcribed to specific crRNA. crRNA guides the Cas protein with endonuclease activity to recognize and degrade specific target sequences [Citation26, Citation27]. Furthermore, in recent years, scientists have discovered that some Cas proteins (e.g. Cas13a) have additional cleavage activity, allowing them to construct a breakthrough new molecular diagnostic method employing CRISPR/Cas technology, which has been hailed as one of seven “disruptive” innovations [Citation28-30]. Zhang Feng's team, for example, used CRISPR/Cas13a combined with RPA technology to identify single nucleic acid molecules. This does meet the criteria for high-sensitivity detection while also meeting the condition for high-specificity detection. Notably, Zhang Feng's research group developed a CRISPR/Cas13a nucleic acid rapid detection technology suitable for lateral flow immunochromatography (LFCA) by modifying biotin and the small-molecule antigen FAM at both ends of the reporter RNA [Citation31], and this technology has been applied to test for SARS-CoV-2 [Citation32]. In a previous study, Cas12a was combined with loop-mediated isothermal amplification (LAMP) technology to achieve HBV DNA detection without requiring specialized equipment [Citation33]. In our study, however, we employed Cas13a in combination with RAA, optimized the procedure, and increased the number of clinical validation samples, particularly for LLV patients.
Long-term LLV infection is linked to the development of liver fibrosis, cirrhosis, and possibly hepatocellular carcinoma [Citation34-36], all of which make it difficult to diagnose and treat HBV infection. According to research, 20%−37.9% of CHB patients have LLV after first-line HBV antiviral therapy, and nearly 30% of patients have low quantities of HBV DNA after 78 weeks of treatment [Citation37]. Therefore, efficient detection of HBV DNA levels is critical for assessing antiviral drug efficacy and enhancing the diagnosis and treatment of LLV patients. To verify our technique, we used 100 LLV patients and 23 antiviral-treated patients (111 dynamic samples). Although an 87% positive coincidence rate was found in the LLV patients, this might be attributed to the inefficiency of HBV DNA extraction and purification. The qPCR assay commonly used in clinical practice extracts HBV DNA from 1 mL of plasma and determines its concentration, whereas we used only 200 µL of plasma during our study, so only one-fifth of the HBV DNA was extracted. Hence, the clinical samples (qPCR quantitative assay results <100 IU/mL) showed instability in the assay, probably related to the HBV DNA extracted by one fifth of it. Furthermore, the specificity was 92.15% and the PPA was 93.75% for the 111 antiviral treatment blood samples since we detected 4 positive samples among those that were negative by qPCR, which also proves the high sensitivity of the method to a certain extent.
We previous developed a novel CRISPR-based assay for the highly sensitive and specific detection of HBV cccDNA, presenting a promising alternative for accurate detection of HBV infection, antiviral therapy evaluation and treatment guidance [Citation38]. The goal of this study is to drive the detection of HBV infection to effectively adaptable for any small to medium-sized laboratory or a field survey. Of course, there are still some deficiencies in our research. For example, not quantitative and limitation of handling number of specimens, comparing to recently advanced high volume through-put and even using autoanalyzer. Moreover, the samples are processed for DNA extraction, RAA, and at last being applied to the test strip, which procedure appears unhandy and difficult to be applied and popularized widely now. However, it has become a reality to complete the lysis of blood samples in 30 s in order to extract and purify DNA [Citation24]. Moreover, a recent study found that the RPA and crRNA-Cas reaction processes may be completed in the same system [Citation39], allowing us to improve the reaction time, streamLine the process and accomplish “one-step” detection. In addition to qPCR, presently RAA isothermal amplification technics are already commercially available at hand, which can fully achieve large-scale promotion and application. Hence, the test strip in this study will fully assist POCT of HBV DNA for clinical use and in low-income countries.
We believe that this rapid and portable HBV DNA detection test strip or other analogous tests will have the ability to rapidly extend the diagnostic screening capacity and coverage of HBV-infected patients, particularly for LLV patients. Furthermore, this study provides a visual and faster alternative to current PCR-based diagnosis for HBV infection. This may simplify the process of validating the efficacy of antiviral therapy in HBV-infected patients and help solve the global public health problem of HBV infection.
Author contributions
FR, XYZ and ZPD designed the paper. YT and ZHF performed and analyzed experiments, and ZHF wrote the paper. LX, YLC, SSC, ZZP, and YG analyzed data and reviewed the paper. HL, and YMM reviewed the paper. FR designed, supervised, and analyzed experimental work and paper. All authors have read and approved the submission of the manuscript.
Supplemental Material
Download PDF (8.4 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Correction Statement
This article has been corrected with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Prevention of mother-to-child transmission of hepatitis B virus: guidelines on antiviral prophylaxis in pregnancy. Geneva: World Health Organization, 2020.
- Tan M, Bhadoria AS, Cui F, et al. Estimating the proportion of people with chronic hepatitis B virus infection eligible for hepatitis B antiviral treatment worldwide: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol. 2021;6(2):106–119. doi:10.1016/S2468-1253(20)30307-1.
- Terrault NA, Lok A, McMahon BJ, et al. Update on prevention, diagnosis, and treatment of chronic hepatitis B: AASLD 2018 hepatitis B guidance. Hepatology. 2018;67(4):1560–1599. doi:10.1002/hep.29800.
- WHO: Global health sector strategy on viral hepatitis 2016-2021. World Health Organization, 2021.
- Bonanzinga S, Onelia F, Jackson K, et al. Multicenter clinical evaluation of alinity m HBV assay performance. J Clin Virol. 2020;129:104514. doi:10.1016/j.jcv.2020.104514.
- Gupta E, Khodare A, Rani N, et al. Performance evaluation of Xpert HBV viral load (VL) assay: point-of-care molecular test to strengthen and decentralize management of chronic hepatitis B (CHB) infection. J Virol Methods. 2021;290:114063. doi:10.1016/j.jviromet.2021.114063.
- Ishizaki A, Bouscaillou J, Luhmann N, et al. Survey of programmatic experiences and challenges in delivery of hepatitis B and C testing in low- and middle-income countries. BMC Infect Dis. 2017;17(Suppl 1):696. doi:10.1186/s12879-017-2767-0.
- Tordrup D, Hutin Y, Stenberg K, et al. Additional resource needs for viral hepatitis elimination through universal health coverage: projections in 67 low-income and middle-income countries, 2016–30. Lancet Glob Health. 2019;7(9):e1180–e1188. doi:10.1016/S2214-109X(19)30272-4.
- Reipold EI, Trianni A, Krakower D, et al. Values, preferences and current hepatitis B and C testing practices in low- and middle-income countries: results of a survey of end users and implementers. BMC Infect Dis. 2017;17(Suppl 1):702), doi:10.1186/s12879-017-2769-y.
- Park HD. Current status of clinical application of point-of-care testing. Arch Pathol Lab Med. 2021;145(2):168–175. doi:10.5858/arpa.2020-0112-RA.
- Boonkaew S, Yakoh A, Chuaypen N, et al. An automated fast-flow/delayed paper-based platform for the simultaneous electrochemical detection of hepatitis B virus and hepatitis C virus core antigen. Biosens Bioelectron. 2021;193:113543), doi:10.1016/j.bios.2021.113543.
- Liu YP, Yao CY. Rapid and quantitative detection of hepatitis B virus. World J Gastroenterol. 2015;21(42):11954–11963.
- Gootenberg JS, Abudayyeh OO, Lee JW, et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356(6336):438–442.
- Ning B, Yu T, Zhang S, et al. A smartphone-read ultrasensitive and quantitative saliva test for COVID-19[J]. Sci Adv, 2021, 7(2). doi:10.1126/sciadv.abe3703.
- Gootenberg JS, Abudayyeh OO, Kellner MJ, et al. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360(6387):439–444. doi:10.1126/science.aaq0179.
- Myhrvold C, Freije CA, Gootenberg JS, et al. Field-deployable viral diagnostics using CRISPR-Cas13[J]. Science. 2018;360(6387):444–448. doi:10.1126/science.aas8836.
- Li H, Dong X, Wang Y, et al. Sensitive and easy-read CRISPR strip for COVID-19 rapid point-of-care testing. CRISPR J. 2021;4(3):392–399. doi:10.1089/crispr.2020.0138.
- Terrault NA, Bzowej NH, Chang KM, et al. AASLD guidelines for treatment of chronic hepatitis B. Hepatology. 2016;63(1):261–283. doi:10.1002/hep.28156.
- Hayer J, Jadeau F, Deléage G, et al. HBVdb: a knowledge database for Hepatitis B Virus. Nucleic Acids Res. 2013;41(Database issue):D566–D570. doi:10.1093/nar/gks1022.
- Sciuto EL, Petralia S, Calabrese G, et al. An integrated biosensor platform for extraction and detection of nucleic acids. Biotechnol Bioeng. 2020;117(5):1554–1561.
- Akram A, Islam S, Munshi SU, et al. Detection of Hepatitis B Virus DNA among Chronic and potential Occult HBV patients in resource-limited settings by Loop-Mediated Isothermal Amplification assay. J Viral Hepat. 2018;25(11):1306–1311. doi:10.1111/jvh.12931.
- Florkowski C, Don-Wauchope A, Gimenez N, et al. Point-of-care testing (POCT) and evidence-based laboratory medicine (EBLM) - does it leverage any advantage in clinical decision making? Crit Rev Clin Lab Sci. 2017;54(7-8):471–494. doi:10.1080/10408363.2017.1399336.
- Nelson PP, Rath BA, Fragkou PC, et al. Current and future point-of-care tests for emerging and New respiratory viruses and future perspectives. Front Cell Infect Microbiol. 2020;10:181.
- Zou Y, Mason MG, Wang Y, et al. Nucleic acid purification from plants, animals, and microbes in under 30 seconds. PLoS Biol. 2017;15(11):e2003916. doi:10.1371/journal.pbio.2003916.
- Pittella-Silva F, Chin YM, Chan HT, et al. Plasma or serum: which is preferable for mutation detection in liquid biopsy? Clin Chem. 2020;66(7):946–957. doi:10.1093/clinchem/hvaa103.
- Pausch P, Al-Shayeb B, Bisom-Rapp E, et al. CRISPR-CasΦ from huge phages is a hypercompact genome editor. Science. 2020;369(6501):333–337. doi:10.1126/science.abb1400.
- Jiang F, Doudna JA. CRISPR-Cas9 structures and mechanisms. Annu Rev Biophys. 2017;46:505–529. doi:10.1146/annurev-biophys-062215-010822.
- Kaminski MM, Abudayyeh OO, Gootenberg JS, et al. CRISPR-based diagnostics. Nat Biomed Eng. 2021;5(7):643–656. doi:10.1038/s41551-021-00760-7.
- Maxmen A. Faster, better, cheaper: the rise of CRISPR in disease detection. Nature. 2019;566(7745):437), doi:10.1038/d41586-019-00601-3.
- Eisenstein M. Seven technologies to watch in 2022. Nature. 2022;601(7894):658–661. doi:10.1038/d41586-022-00163-x.
- Freije CA, Myhrvold C, Boehm CK, et al. Programmable inhibition and detection of RNA viruses using Cas13. Mol Cell. 2019;76(5):826–837.e11. doi:10.1016/j.molcel.2019.09.013.
- Broughton JP, Deng X, Yu G, et al. CRISPR–Cas12-based detection of SARS-CoV-2. Nat Biotechnol. 2020;38(7):870–874.
- Ding R, Long J, Yuan M, et al. Crispr/Cas12-based ultra-sensitive and specific point-of-care detection of HBV[J]. Int J Mol Sci, 2021, 22(9). doi:10.3390/ijms22094842.
- Sinn DH, Lee J, Goo J, et al. Hepatocellular carcinoma risk in chronic hepatitis B virus-infected compensated cirrhosis patients with low viral load. Hepatology. 2015;62(3):694–701. doi:10.1002/hep.27889.
- Kim JH, Sinn DH, Kang W, et al. Low-level viremia and the increased risk of hepatocellular carcinoma in patients receiving entecavir treatment. Hepatology. 2017;66(2):335–343. doi:10.1002/hep.28916.
- Ogawa E, Nomura H, Nakamuta M, et al. Tenofovir alafenamide after switching from entecavir or nucleos(t)ide combination therapy for patients with chronic hepatitis B. Liver Int. 2020;40(7):1578–1589. doi:10.1111/liv.14482.
- Sun Y, Wu X, Zhou J, et al. Persistent Low level of hepatitis B virus promotes fibrosis progression during therapy. Clin Gastroenterol Hepatol. 2020;18(11):2582–2591.e6. doi:10.1016/j.cgh.2020.03.001.
- Zhang X, Tian Y, Xu L, et al. CRISPR/Cas13-assisted hepatitis B virus covalently closed circular DNA detection. Hepatol Int. 2022;16(2):306–315. doi:10.1007/s12072-022-10311-0.
- Wang Y, Ke Y, Liu W, et al. A One-Pot toolbox based on Cas12a/crRNA enables rapid foodborne pathogen detection at attomolar level. ACS Sens. 2020;5(5):1427–1435. doi:10.1021/acssensors.0c00320.

