ABSTRACT
Since May 2022, human mpox cases have increased unexpectedly in non-endemic countries. The first imported case of human mpox in Hong Kong was reported in September 2022. Here we report the isolation and identification of MPXV from the vesicle swabs of this patient. In this research, the vesicle swabs were inoculated in Vero and Vero E6 cells. In addition to observing cytopathic effects (CPEs) in Vero or Vero E6 cells, the isolated virus was identified as mpox virus (MPXV) using quantitative Real-Time PCR (RT–PCR), transmission electron microscopy, and high-throughput sequencing. The experiment also assessed the cross-protective efficacy of sera from the smallpox vaccinated population and preliminarily assessed the inhibitory effect of anti-smallpox virus drugs against MPXV. CPEs can be observed on Vero E6 cells at 24 h and Vero cells at 48 h. The virus particles could be observed by transmission electron microscope, showing typical orthopoxvirus morphology. In addition, F3L and ATI genes which from MPXV A39R, B2R, HA genes which from orthopoxvirus were confirmed by conventional PCR and Sanger sequencing. The next generation sequencing (NGS) suggests that the MPXV strain belongs to B.1 branch of the West African linage, and has a high identity with the sequence of the 2022 ongoing outbreak. PRNT50 results showed that 26.7% of sera from individuals born before 1981 who had been immunized with smallpox were positive, but no MPXV-neutralizing antibodies were found in sera from individuals born later. All four anti-smallpox virus drugs evaluated demonstrated inhibition of mpox virus.
GRAPHICAL ABSTRACT
Introduction
Mpox virus (MPXV) is a member of the Orthopoxvirus (OPXV) genus of the Poxviridae family, which is closely related to the infamous variola (smallpox) virus, causing a febrile rash illness in humans similar to but milder than smallpox [Citation1]. While smallpox was eradicated in 1980, mpox continues to occur in countries of central and west Africa. Mpox is an illness caused by the mpox virus. In the twentieth century, human mpox had been mostly a rare zoonotic disease confined to forested areas in West and Central Africa [Citation1]. Since May 2022, mpox cases have also been reported in some non-endemic countries and then MPXV rapidly spread in Europe and the United States among individuals who had not travelled to endemic areas. Mpox was declared a Public Health Emergency of International Concern (PHEIC) by the World Health Organization (WHO) Director General Tedros Adhanom Ghebreyesus on 23 July 2022 [Citation2]. As of 16 November 2022, more than 79,641 cases across 110 countries have been reported since January 2022 [Citation3]. A mpox case was first imported in Hong Kong on 6 September 2022, which presented with infectious mononucleosis-like syndrome [Citation4], followed by the first case of mpox in the mainland of China on 26 September 2022 in Chongqing [Citation5].
Vaccination against smallpox generally provides some protection against mpox [Citation6]. Since the cessation of smallpox vaccination in 1980, herd immunity has steadily declined. In July 2018, the Food and Drug Administration (FDA) approved Tecovirimat for the treatment of smallpox. In January 2022, the European Medicines Agency (EMA) approved the launch of Tecovirimat for the treatment of cowpox, smallpox and mpox. According to the provisional clinical guidelines for mpox treatment issued by the centres for disease control and prevention (CDC) of the United States, the antiviral drugs currently developed for smallpox patients may be beneficial for mpox, including Brincidofovir, Cidofovir, vaccinia immune globulin (VIG), etc. In addition, there are also some broad-spectrum anti DNA virus drugs that can be used for the treatment of mpox, such as Idoxyuridine. In order to prevent the spread of mpox virus, the most urgent task is to determine the cross immune protection effect of smallpox vaccine which was used in China as vaccinia virus (VACV) Tiantan strain (VTT) against mpox virus, and find effective drugs to treat MPXV infections.
In this study, we report the isolation and identification of the MPXV which was named WIBP-MPXV-001 derived from a swab taken from a typical pox lesion of an MPXV-positive Hong Kong patient by inoculating Vero and Vero E6 cells. Using next-generation sequencing (NGS), the infected cells were analysed to identify potential aetiological agents and to identify genetic differences that drive the epidemiological characteristics. In addition, we evaluated the neutralizing antibody level in serum samples from participants based on year of birth, and divided over decades ≤1981 (N = 15) and >1981 (N = 15) (smallpox vaccination of the general population was stopped in 1981 in China), and the effects of drugs against mpox virus.
Materials and methods
Virus isolation from clinical specimens
The clinical specimen was vesicle swabs infiltrated with viral transport medium. The specimens were diluted with antibiotics (penicillin/streptomycin) at a 1:4 ratio, and then incubated with equivalent volumes of Dulbecco’s modified Eagle’s Medium (DMEM, Gibco, USA) supplemented with 2% NBS and 50 units/ml Penicillin Streptomycin (Gibco, USA) for 30 min at 4°C. After centrifugation at 3000 rpm for 5 min, the supernatant was inoculated into Vero and Vero E6 cells. All virus isolation procedures were performed in a biosafety level 3 facility.
RT–PCR for the detection of MPXV
Whenever commercial kits were used, the manufacturing instructions were followed without modification. Viral DNA was extracted from 200 μL samples using the DAAN Nucleic Acid Extraction or Purification Kit (DAAN Gene, China). Due to the rarity of the first-generation samples, 20 μL of viral culture supernatant was absorbed and supplemented with 180 μL DMEM for nucleic acid extraction. RT–PCR was performed on the QuantStudio 5 real-time PCR system (Thermo Fisher Scientific, USA) with mpox virus nucleic acid detection kit targeting F3L fragment (DA AN gene, China).
Transmission electron microscopy
Vero E6 cells were infected with the supernatant of the treated swab and the supernatant of infected cells was collected at 72 h post infection (hpi) and mixed with 8% paraformaldehyde at the ratio 1:1 for more than one hour. Then 20 μL fixed sample was adsorbed to a Formvar Stabilized 230-mesh with Carbon Support Film for 5 min at room temperature. The copper grid was soaked with 20 μL 3% phosphotungstic acid and allowed to dry for 5 min at room temperature. After that, remove the copper mesh and use filter paper to absorb excess liquid drops on its edge. The staining grid was observed with a transmission electron microscope (Talos L120C TEM, ThermoFisher, Germany) at an acceleration voltage of 120 kV.
Conventional PCR for the detection of MPXV
Conventional polymerase chain reaction (PCR) was performed with nucleic acids obtained from the extraction described above and selected primer sets. The sequences of the primer sets for conventional PCR (ProFlex PCR system, Applied Biosystems, USA) were as follows: F3L-forward primer, 5’-CATCTATTATAGCATCAGCATCAGA-3’, F3L-reverse primer, 5’-GATACTCCTCCTCGTTGGTCTAC-3’ for the F3L gene; A39R-forward primer, 5’-TGGGATAACGAATCCAATGTCA-3’, A39R-reverse primer, 5’-GCGTGCTTCCAGCAACACT-3’ for the A39R gene [Citation7]; 1995MPV-forward primer, 5’-CTGATAATGTAGAAGAC-3’; 1995MPV-reverse primer, 5’-TTGTATTTACGTGGGTG-3’ for the B2R gene [Citation8]; 1997MPV-forward primer, 5’-AATACAAGGAGGATCT-3’; 1997MPV-reverse primer, 5’-CTTAACTTTTTCTTTCTC-3’ for the ATI gene; HAOUT-forward primer, 5’-CCATTGGAAAAAACACAGTAC-3’; HAOUT-reverse primer, 5’-CCAAATATATTCCCATAGTC-3’ for the HA gene [Citation9]. The PCR conditions for the A39R gene were as follows: 98°C for 2 min, 35 cycles of 98°C for 10 s, 58°C for 10 s, and 72°C for 10 s, followed by 72°C for 10 min. The conventional PCR conditions for the other genes were as follows: 98°C for 2 min, 35 cycles of 98°C for 10 s, 55°C for 10 s, and 72°C for 10 s, followed by 72°C for 10 min. The PCR products were sent to Sangon Biotech (Shanghai, China) for sequencing analysis with a 3730xl DNA analyzer (Applied Biosystems, USA), sequence reactions using the F3L, A39R, 1995MPV, 1997MPV, and HAOUT primers. A nucleotide blast was carried out with the sequencing results to confirm whether the sequences matched those of MPXV.
High-throughput sequencing, pathogen screening and genome assembly, phylogenetic analysis
Total DNA purification was performed using 200 μL of the supernatant of virus cultures and the FastPure® Viral DNA/RNA Mini Kit (Vazyme, China) used for de novo sequencing and FastPure® Microbiome DNA Isolation Kit (Vazyme, China) used for metagenomics sequencing, according to the manufacturer’s instructions and eluting in 50 μL of elution buffer. DNA was quantified using fluorimetry with the Qubit dsDNA High Sensitivity Assay (Invitrogen, USA) on the Qubit 4.0 instrument (Life Technologies, USA). VAHTS Universal Plus DNA Library Prep Kit for MGI (Vazyme, China) was prepared using 10 ng of extracted DNA. Libraries were prepared with an input of 50 ng per sample, pooled in an equimolar way. The library was sequenced on the BGI MGISEQ2000 platform using paired-end sequencing, with a read length of 100 nucleotides (NT). The raw NGS reads were first processed by Cutadapt (v.1.18) with a minimum read length of 30 base pairs. Alignment process in which the reads are then aligned to a reference genome of MPXV-UK_P2 (GenBank accession no. MT903344.1) provided in the ref.fa file with BWA-MEM. The resulting SAM and BAM files are processed including sorting and filling in Samtools and the low coverage regions are masked with BEDtools. After masking a variant call is done with freebayes before computing the consensus sequence via BCFtools of Samtools. Routine sequence management and analysis were carried out using DNAStar. The sequence alignment of complete genome sequences was performed using MAFFT with default parameters.
PCR and Sanger sequencing were performed to confirm the NGS result. The sequences of the primers for PCR were in supplement 9. The PCR conditions were as follows: 98°C for 2 min, 35 cycles of 98°C for 10 s, 55°C for 10 s, and 72°C for 10 s, followed by 72°C for 10 min.
The tree was constructed by the maximum likelihood method with IQTree2. Diamond software was used to compare the gene sequences to the Uniprot database, then the mapping relationship between the database Uniprot Id and Gene Ontology (GO) Id was used to obtain the GO annotation results.
Detection of MPXV IgG antibodies by ELISA
Serum samples were 1:10 diluted in standard diluent. MPXV IgG antibodies were detected using Epoch Microplate Reader (Agilent BioTek, USA) with Human Anti-Mpox Virus (MPXV) IgG ELISA Kit (A29L) (Antibody System, France). The IgG concentration was given a value of 10 when the value is below the detection limit.
Detection of MPXV neutralizing antibodies by Plaque Reduction Neutralization Tests (PRNT)
Vero E6 cells were seeded one day prior to the experiment in 12-well plates (Corning, Germany) at a density of 4∼5 × 105 cells per well. Serum samples were 1:16 diluted, followed by a 4-fold serial dilution in DMEM supplemented with 2% NBS, 50 units/ml Penicillin Streptomycin (Gibco, USA). Each serum sample was incubated with 150 plaque-forming units of virus (PFU) for 1 h at 37°C. The virus-serum mixtures were added onto pre-formed Vero E6 cell monolayers and incubated for 1 h at 37°C in 5% CO2 incubator. Then the supernatant was removed and the cell monolayers were covered with methylcellulose overlay (final concentration: DMEM with 0.9% Methylcellulose, 2% NBS and 50 U/mL penicillin–streptomycin). After 4 days, the plates were fixed with an equal volume of 8% paraformaldehyde for more than 1 h, the cell monolayers were stained with 0.5% crystal violet, and then the plaques were counted and photographed. Neutralizing antibody titres were defined as the highest serum dilution that resulted in >50% (PRNT50) in the number of virus plaques. The PRNT50 was given a value of 10 when no neutralization was observed.
Drug inhibition test
Vero E6 cell monolayers were seeded in 96-well plates (2 × 104 cells per well). Dose–response curves were generated by measuring virus-induced cytopathic effects in the presence of a range of compound concentrations. Four drugs were selected for this drug inhibition assay, namely Tecovirimat (AbMole,USA), Cidofovir (MedChemExpress,USA), Brincidofovir (MedChemExpress,USA) and Idoxuridine (MedChemExpress,USA). Eight compound concentrations (5, 1.5, 0.5, 0.15, 0.05, 0.015, 0.005 and 0.0015 μM) were used to generate curves of Cidovir and Idoxuridine, 12 compound concentrations, with the maximum concentration of 200 μM and continue to six-fold dilution were used to generate curves of Tecovirimat and Brincidofovir, which is suitable for calculating EC50 from virus copy number by RT–PCR. Eleven compound concentrations, with a maximum concentration of 200 μM and continue to triple dilution were used to generate curves suitable for calculating the cytotoxic concentration of compound that inhibited cell viability by 50% (CC50) by CCK-8 method. Compound dilutions were prepared in dimethyl sulfoxide (DMSO, Sinopharm Chemical Reagent, China) prior to addition to the cell culture medium. This dose of virus was 0.005 PFU/cell. The results were collected on the fifth day after infection. EC50s were calculated by fitting the data to a Log (Inhibition) vs. Normalized Response model (variable slope, nonlinear regression model) to generate a dose–response curve using GraphPad Prism 8.0.2.
Statistical analysis
Data of MPXV IgG and neutralizing antibodies by ELISA and PRNT between different groups were tested for statistical normality and the results were used to apply one-way ANOVA tests and t-test analysis. P values <0.05 were considered significant. All statistical analyses were performed using GraphPad Prism 8.
Results and discussion
Virus isolation and identification
In the inoculation of vesicle swabs, CPEs could be observed on Vero E6 cells after 24 hpi. While it could be observed at 48 hpi on Vero cells. After 3 days, CPEs and detaching of cells were visible on the entire area of the monolayers ((A)). The virus was clonally purified through plaque purification and that culture supernatant and infected cells were obtained after infecting the cells with the plaque-purified virus. The culture supernatant and cell lysates were used for examinations such as RT–PCR, electron microscopy, and sequenced as described in the following sections.
Figure 1. Isolation and identification of WIBP-MPXV-001. (A) Cytopathic effects of Vero and Vero E6 cells at 24, 48, and 72 hpi, with cell control. (B), (C) RT-PCR amplification plot and cycle threshold of culture suspension and infected cells. (D) Electron microscopy of WIBP-MPXV-001. (E) Conventional polymerase chain reaction products of WIBP-MPXV-001. M, size maker; lane 1,blank; lane 2, B2R; lane 3, A39R; lane 4, ATI; lane 5, F3L; lane 6, HA. (F) Conventional polymerase chain reaction products of 43 InDels and 8 SNP mutations of WIBP-MPXV-001 compared with the mpox/PT0001/2022. (G) Metagenomics analysis of next-generation sequencing of cells infected with the virus.
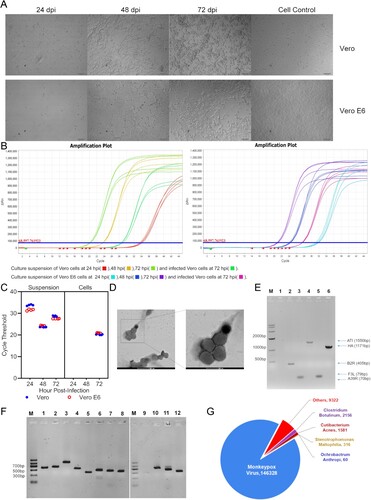
RT–PCR results on 24, 48 and 72 hpi culture supernatants and 72 hpi infected cells all showed that virus copy numbers were detected, and the copy number of 72 hpi Vero cells was higher than that of Vero E6 cells ((B, C)).
Transmission electron microscopy revealed that WIBP-MPXV-001 viruses were present in Vero E6 cells. In this study, mulberry-shaped particles with diameters between 200 and 250 nm were observed, which are typical of MPXV ((D)).
The isolated virus was confirmed as MPXV by PCR assays ((E)). Conventional PCR assays with F3L primers for the F3L gene, A39R primers for the A39R gene, 1995MPV primers for the B2R gene, 1997MPV primers for the ATI gene, and HAOUT primers for the HA gene were performed, followed by Sanger sequencing. As shown in (E), the F3L gene is 79 bp, the A39R gene is 70 bp, the B2R gene is 405 bp, the ATI gene is 1550 bp and the HA gene is 1171 bp. The Sanger sequencing results showed that sequences of those genes were >98% identical to previously reported human MPXVs (OP555662.1;OP555661.1;OP555659.1). In addition, conventional PCR detecting the ATI genes of the mpox viruses, was designed to distinguish 2 clades of MPXV-clade I (Congo-Basin clade, 450 bp) and clade II (West African, 1550 bp). The size of the PCR product of MPXV was 1550 bp, indicating that the virus belonged to the West African clade. After, confirming the virus as MPXV, we named the virus WIBP-MPXV-001.
Genome sequencing, prediction and mutant annotation
The infected cell lysates were analysed by metagenomics analysis using next-generation sequencing to identify potential aetiological agents. Of the 9677303 total reads – of which 159763 total reads were retained after filtering of reads from the Vero E6 genome – 146328 (91.59%) of the sequences matched the sequence of MPXV ((G)). By de novo assembly, the sequencing produced generated a total of 106327280 reads, which covered 100% of the viral genome and produced a genome map of 197160-base-pairs, and 33.01% of which were G:C ((C)).
Figure 2. Sequence analysis of WIBP-MPXV-001 isolated in Vero E6 cells. (A) Phylogenetic tree of WIBP-MPXV-001. (B) NR species distribution map of WIBP-MPXV-001. (C) Genome circle map of WIBP-MPXV-001. From the outside to the inside, the first circle is genome sequence, the second and third circle are G:C content and GC skew curve respectively, the fourth circle is sequencing depth and coverage information, and the fifth circle shows CDS and non-coding regions. (D) Go analysis of WIBP-MPXV-001.
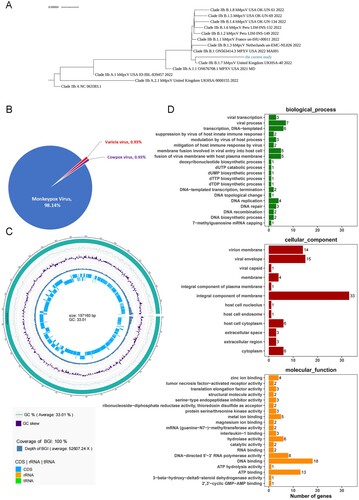
More extensive comparative analyses were carried out between WIBP-MPXV-001 with West African SL-V70 (NCBI accession number AY741551.1), Congo basin ZAI-96 (NCBI accession number AF380138.1) and the first outbreak-related MPXV genome sequence of 2022 (mpox/PT0001/2022) [Citation10], as ZAI-96 and SL-V70 are classic Congo and West African mpox strains, respectively. WIBP-MPXV-001 showed 43 InDels and 8 SNP mutations compared with the mpox/PT0001/2022 (Supplement Tables 1–2) and several unique nucleotide mutations were detected in our strain including C25653 T, C57182 T, C142795 T, C149135 T, G150704A, G173289 T, G186733A, C188433 T, and InDels at nucleotide position 617, 15299, 133087–133094, 133174, 1367551, 173268-173286, 173291. Sanger sequencing were performed and the results were consistent with NGS sequencing,except SNP nucleotide position at 173289 and InDels at nucleotide position 617, 173268∼173286 and173291, which failed after multiple sequencing attempts due to the polyA structure of the fragment ((F), Supplement 10). 471 InDels and 402 SNP mutations compared with the SL-V70 (Supplement Tables 3–4), 875 InDel and 959 SNP mutations with ZAI-96 (Supplement Tables 5–6). The results showed that some mutations are in the non-coding region, which will not cause amino acid mutations, while some mutations are in the coding region, which may cause amino acid changes in the protein, and further studies are needed to analyse the active sites of proteins. The phylogenetic tree showed that the sequence belonged to B.1 and was related to sequences from the 2022 sequences from many countries, especially sequences from the UK ((A)).
NR database analysis showed WIBP-MPXV-001 have 98.14%, 0.93% and 0.93% nucleotide base percentage similarity with mpox, cowpox and smallpox, respectively ((B)). To further analysis the genome, Prokka (Version: 1.14.6, Seemann 2014) software was used to predict coding genes of the assembled genome. A total of 216 genes were predicted, of which 215 were CDS regions encoding proteins and one was encoding miscRNA (Supplement Table 7). Then Reference Sequences (Refseq) and databases were used to annotate the genes of WIBP-MPXV-001 (Supplement Table 8, ). The results showed that 169 genes of WIBP-MPXV-001 were highly homologous to mpox and 37 genes were highly homologous to genes of other viruses of orthopoxaceae, such as vaccinia, smallpox and cowpox. Proteins including Cytokine response-modifying proteins, dUTPase, membrane proteins, late transcription elongation factors, Resolvase, Golgi anti-apoptotic protein, Interleukin-1-binding protein and interferon alpha/beta receptor, etc. were predicted and annotated.
GO analysis was used to make functional annotations of the predicted genes ((D)). It showed that the functional annotation of WIBP-MPXV-001 was mainly concentrated on biological processes such as viral transcription, membrane fusion involved in viral entry into host cells, dNTP synthesis, DNA replication and repair, mitigation of host immune response and suppression of host innate immune response. Cellular component analysis revealed that these proteins of WIBP-MPXV-001 mainly expressed in virion membrane, envelope, capsid, integral component of membrane and nucleolus, endosome, cytoplasm of host cell and extracellular region. Molecular function analysis showed that these proteins have ion activity, transcription elongation activity, Interleukin-1-binding activity.
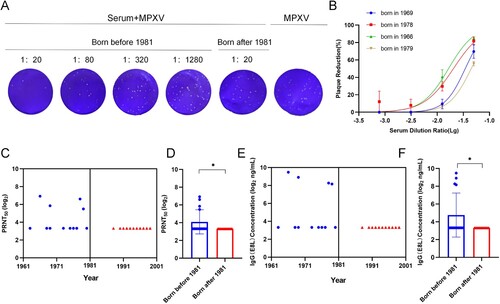
Detection of MPXV IgG and neutralizing antibodies by ELISA and PRNT
Thirty seras from 30 individuals who were immunized (N = 15, of which N = 15 sera from individuals born ≤ 1981) and non-immunized with smallpox vaccine (N = 15, of which N = 15 sera from individuals born > 1981) were randomly tested to assess the presence of antibodies capable of binding MPXV. Only 26.7% of sera collected from individuals born ≤ 1981 were positive through MPXV IgG Elisa kit ((E)), which is consistent with MPXV-neutralizing antibodies ((A–C)). Sera not capable of neutralizing MPXV were also negative for MPXV IgG antibodies in the ELISA. Binding and neutralizing antibodies have a significant increase (neutralizing antibodies: p = 0.0452; binding antibodies: p = 0.0417) in groups born before 1981 than after 1981 ((D,F)).
Figure 3. Detection of MPXV IgG and neutralizing antibodies by ELISA and PRNT. (A) Plaque reduction assay of serum against MPXV. (B) The inhibitory curve for positive sera. (C) Neutralizing activity of sera from individuals (N = 30) who were born before 1981 and after 1981. (D) Geometric mean titre (GMT) of neutralizing antibodies against MPXV. (E) MPXV IgG antibodies of sera from individuals (N = 30) who were born before 1981 and after 1981 by ELISA. (F) GMT of IgG antibodies by ELISA. There were two people born in the same year such as 1968, 1969, 1977, 1992, 1996, 1997, and 1999 respectively, which resulted in 23 data points shown (7 data points less) in C and E. Data shown are geometric mean titres. Error bars represent geometric standard deviations. Data were analysed by the one-way ANOVA tests and t-test analysis. P < 0.05 was considered statistically significant. *P < 0.05.
Drug inhibition test
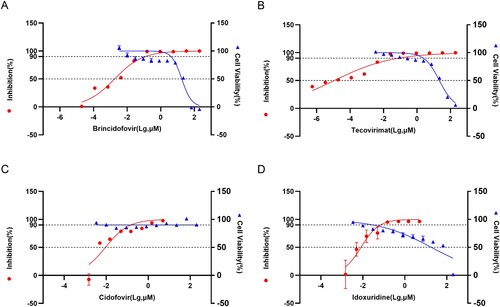
When the virus dose is 0.005 PFU/cell, the inhibitory effect of Brincidofovir (EC50 = 0.00179 μM) and Tecovirimat (EC50 = 1.137 × 10−5 μM) is better than Cidovir (EC50 = 0.00916 μM) and Idoxuridine (EC50 = 0.00811 μM). The cytotoxicity test results of the drug showed that the CC50 of Tecovinimat was 21.95 μM, the CC50 of Idoxuridine was 18.33 μM, the CC50 of Brincidofovir was 17.87 μM, and the CC50 of Cidovir > 200 μM ().
Figure 4. Drug Inhibition Test. (A) Cytotoxicity of Brincidofovir and its Inhibition efficiency on WIBP-MPXV-001 (B) Cytotoxicity of Tecovirimat and its Inhibition efficiency on WIBP-MPXV-001 (C) Cytotoxicity of Cidofovir and its Inhibition efficiency on WIBP-MPXV-001 (D) Cytotoxicity of Idoxuridine and its Inhibition efficiency on WIBP-MPXV-001.
Discussion
WIBP-MPXV-001 was successfully isolated from a Chinese patient. RT–PCR results indicated that the virus underwent proliferation, which was also reflected in CPE. Using PCR, sequencing and electron microscopy techniques, the virus isolated in Vero or Vero E6 cells was identified as MPXV.
Sequence alignment and phylogenetic tree analysis showed that WIBP-MPXV-001 belonged to the West African Clade and was highly homologous to the 2022 outbreak. Preliminary genomic sequencing studies have implicated that the 2022 outbreak is caused by lineage B.1 of West African clade [Citation11].
MPXV is a DNA virus and is generally more stable than RNA viruses like influenza and SARS-CoV-2 [Citation12]. The earliest human case of mpox was in 1970 in Zaire [Citation13]. The clinical presentation of mpox does not seem to have changed very much over the past 40 years [Citation14]. However, WIBP-MPXV-001 showed the presence of mutations compared with classical MPXV strains such as the SL-V70 and ZAI-96. It’s interesting that WIBP-MPXV-001 still had 43 InDels and 8 SNP mutations when compared to the first sequence of the 2022 outbreak. Narendra Kumar et al. reported that the isolates from the 2022 outbreak shared 40 mutations that distinguish it from its closest variant [Citation15]. Previous research also reported that MPXV had been circulating in humans and had a mutation rate around 10 times higher than the virus’s standard mutation rate since 2017. The virus isolated from the 2022 outbreak of MPXV seems to have more mutations and has shown a higher than expected rate of genomic variance [Citation11]. While MPXV appears to have mutated due to its ability to transmit among people, it is unclear what caused the accelerated evolution of mutants and whether mutants affect proteins.
In the twentieth century, human mpox was mostly a rare zoonotic disease confined to forested areas in West and Central Africa [Citation1]. Since May 2022, mpox cases have also been reported in some non-endemic countries and then MPXV rapidly spread in Europe and the United States among individuals who had not travelled to endemic areas. Nevertheless, it remains unclear how transmission has changed. WIBP-MPXV-001 encodes 216 genes, many of which do not have well-defined functions [Citation16]. Genome prediction result showed that genes inclouding cytokine response-modifying proteins, interleukin-1-binding protein and interferon alpha/beta receptor, etc. were predicted and annotated. GO analysis showed that some proteins of WIBP-MPXV-001 have interleukin-1-binding activity and participate in biological processes such as mitigation of host immune response and suppression of host innate immune response et al. In reported cases of human MPXV infection, numerous cytokines are elevated following infection, these include IL-1β, IL-1RA, CCL2 and CCL5 [Citation17]. Immune evasion strategies studies have showed that MPXV can interfere with interferon signalling by blocking IFNα/β binding or suppressing IFNα/β production. In addition, MPXV secretes proteins that can target key inflammatory molecules such as TNF, IFNγ, IL-1β [Citation18].
Induction of neutralizing antibodies by smallpox vaccines is considered to be an important immunologic factor in protection against smallpox [Citation19]. Several smallpox vaccines have been shown to be effective in preventing MPXV infection [Citation20]. Vaccinia virus (VACV) Tiantan strain (VTT) has played a significant role in eradicating smallpox in China in the past [Citation21]. Currently, the spread of MPXV has become a global concern. In a previous study, VACV-based vaccines for smallpox including MVA [Citation20], LC16m8 [Citation22], ACAM2000 [Citation23], and DryVAX [Citation24] showed 85% protection against mpox [Citation25]. It was reported that two commercial vaccines for smallpox, JYNNEOS and ACAM2000 could potentially to elicit highly cross-reactive immunity [Citation26]. Nevertheless, it remains unclear whether the Chinese smallpox vaccine VTT will be as effective as MVA-BN or ACAM2000. Lei Yang et al. reported that VTT-elicited antibodies could effectively cross-react with MPXV protective antigens, implying that this traditional smallpox vaccine could still be valuable in MPXV prevention [Citation27]. However, more experiments using live MPXV are required to determine these neutralizing antibody titres. Here, we measured MPXV-binding and neutralizing antibodies of historic smallpox-vaccinated individuals. We only could detect MPXV-reactive antibodies in 4/15 individuals born before 1981, and the group born before 1981 had significant increases in binding and neutralizing antibodies, which may be due to historic smallpox vaccination. Significantly, there are still undetected mpox binding and neutralizing activity of sera in 11/15 individuals born before 1981. It may be related to smallpox vaccination rates, vaccine cross-reactivity, and vaccine durability. Similar findings have been reported in earlier studies, it is possible to detect neutralizing antibodies in pre-immune individuals by using serology [Citation28]. Additionally, these also support earlier assertions about the longevity of antibodies induced by smallpox-vaccinated [Citation29, Citation30].
Tecovirimat targets viral F13L phospholipase as a viral efflux inhibitor and blocks the formation of OPV envelope forms, thereby effectively inhibiting viral transmission in vitro and in vivo [Citation31]. Cidofovir is a nucleoside analogue that selectively inhibits viral DNA polymerase and reduces the replication of smallpox virus in vitro [Citation32]. However, Cidofovir can cause significant nephrotoxicity. Its lipid analogue, Brincidofovir, is available by the oral route and no nephrotoxicity has been reported [Citation33]. In addition, Idoxuridine and its various analogues have been reported to inhibit DNA replication, either directly or indirectly, to achieve antiviral effects [Citation34]. In this study, the EC50 results of the combined evaluation of these four drugs using WIBP-MPXV-001 showed that Tecovirimat exhibited superior antiviral effects against mpox virus. The other three anti-smallpox drugs were also effective against mpox virus, which has important implications for the development of new drugs and implies that the previous idea of developing drugs based on orthopox virus is also applicable to the new mutant strains of mpox virus in 2022.
Inherently, the present study has some limitations. WIBP-MPXV-001 showed the presence of mutations compared with classical MPXV strains such as SL-V70 and ZAI-96. However, it is unclear the reasons for the accelerated evolutionary mutations and whether the mutants affect protein function, as well as whether these changes lead to a change in transmission mode. The number of sera analysed from vaccine recipients is low. As we did not have access to historic vaccination records, we could not confirm that people born prior to 1981 have been vaccinated against smallpox, or how many shots they received. This study was not ascertain the efficacy of the vaccine and drugs in an animal infection model. We analysed the efficacy only on cells. At this moment it is unclear what the relatively MPXV-neutralizing titres mean for protection against disease, severity of symptoms, and transmissibility. Future work with more individuals and longer survey periods will help to determine the above-mentioned.
Ethics declarations
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2008. Informed consent was obtained from all patients for being included in the study.
Acknowledgements
The authors thank Public Health Laboratory Services Branch, in Hong Kong, China for providing the vesicle swabs of a mpox-infectious case and P. Zhang from the WIV core facility centre for their help with producing transmission electron microscopy micrographs.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Correction Statement
This article has been corrected with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Xiang Y, White A. Monkeypox virus emerges from the shadow of its more infamous cousin: family biology matters. Emerg Microbes Infect. 2022;11(1):1768–1777.
- Wenham C, Eccleston-Turner M. Monkeypox as a PHEIC: implications for global health governance. Lancet. 2022 Dec 17;1400(10369):2169–2171.
- Organization WH. Monkeypox Outbreak Global [map]. 2022. Available from: https://worldhealthorg.shinyapps.io/mpx_global/
- Chiu KH, Wong SC, Tam AR, et al. The first case of monkeypox in Hong Kong presenting as infectious mononucleosis-like syndrome. Emerg Microbes Infect. 2023;12(1):2146910
- Zhao H, Wang W. The first imported case of monkeypox in the mainland of China – Chongqing Municipality, People’s Republic of China. China CDC Wkly [Internet]. 2022 Sep 16; 4(38): 853–854.
- Fine PE, Jezek Z. The transmission potential of monkeypox virus in human populations. Int J Epidemiol. 1988;17(3):643–650.
- Jang YR, Lee M. The first case of monkeypox in the Republic of Korea. J Korean Med Sci. 2022;37(27):e224.
- Ropp SL, Jin Q. PCR strategy for identification and differentiation of small pox and other orthopoxviruses. J Clin Microbiol. 1995;33(8):2069–e276.
- Damaso CR, Esposito JJ. An emergent poxvirus from humans and cattle in Rio de Janeiro State: cantagalo virus may derive from Brazilian smallpox vaccine. Virology. 2000;277(2):439–449.
- Joana Isidro1 VB. First draft genome sequence of Monkeypox virus associated with the suspected multi-country outbreak (confirmed case in Portugal). 2022 May. Available from: https://virological.org/t/first-draft-genome-sequence-of-monkeypox-virus-associated-with-the-suspected-multi-country-outbreak-may-2022-confirmed-case-in-portugal/799.
- Isidro J, Borges V. Phylogenomic characterization and signs of microevolution in the 2022 multi-country outbreak of monkeypox virus. Nat Med. 2022;28(8):1569–1572.
- Sanjuán R, Domingo-Calap P. Mechanisms of viral mutation. Cell Mol Life Sci. 2016;73(23):4433–4448.
- Ladnyj ID, Ziegler P, Kima E. A human infection caused by monkeypox virus in Basankusu Territory, Democratic Republic of the Congo. Bull World Health Organ. 1972;46(5):593–597.
- Burki T. Investigating monkeypox. Lancet. 2022;399(10343):2254–2255.
- Kumar N, Acharya A, Gendelman HE. The 2022 outbreak and the pathobiology of the monkeypox virus. J Autoimmun. 2022;131:102855.
- Shchelkunov SN, Totmenin AV. Analysis of the monkeypox virus genome. Virology. 2002;297(2):172–102194.
- Johnston SC. Cytokine modulation correlates with severity of monkeypox disease in humans. J Clin Virol. 2015;63:42–45.
- Lum FM, Torres-Ruesta A. Monkeypox: disease epidemiology, host immunity and clinical interventions. Nat Rev Immunol. 2022;22(10):597–613.
- Mack TM, Noble J, Thomas DB. A prospective study of serum antibody and protection against smallpox. Am J Trop Med Hyg. 1972;21(2):214–218.
- Earl PL, Americo JL. Immunogenicity of a highly attenuated MVA smallpox vaccine and protection against monkeypox. Nature. 2004;428(6979):182–185.
- Fang Q, Yang L, Zhu W, et al. Host range, growth property, and virulence of the smallpox vaccine: vaccinia virus Tian Tan strain. Virology. 2005;335(2):242–251.
- Saijo M, Ami Y, Yuriko S, et al. LC16m8, a highly attenuated vaccinia virus vaccine lacking expression of the membrane protein B5R, protects monkeys from monkeypox. J Virol. 2006;80(11):5179–51788.
- Golden JW, Josleyn M. Side-by-side comparison of gene-based smallpox vaccine with MVA in nonhuman primates. PLoS One. 2012;7(7):e42353.
- Edghill-Smith Y, Golding H, Manischewitz J, et al. Smallpox vaccine-induced antibodies are necessary and sufficient for protection against monkeypox virus. Nat Med. 2005;11(7):740–747.
- Organization WH. Monkeypox. Available from: https://www.who.int/news-room/fact-sheets/detail/monkeypox
- Ahmed SF, Sohail MS. Vaccinia-virus-based vaccines are expected to elicit highly cross-reactive immunity to the 2022 monkeypox virus. Viruses. 2022;14:9.
- Yang L, Chen Y, Li S, et al. Immunization of mice with vaccinia virus Tiantan strain yields antibodies cross-reactive with protective antigens of monkeypox virus. Virol Sin. 2023 Feb;38(1):162–164.
- Zaeck LM, Lamers MM, Verstrepen BE, et al. Low levels of monkeypox virus-neutralizing antibodies after MVA-BN vaccination in healthy individuals. Nat Med. 2023 Jan;29(1):270–278.
- Hammarlund E, Lewis MW, Carter SV. Multiple diagnostic techniques identify previously vaccinated individuals with protective immunity against monkeypox. Nat Med. 2005;11(9):1005–1011.
- Taub DD, et al. Immunity from smallpox vaccine persists for decades: a longitudinal study. Am J Med. 2008;121(12):1058–1064.
- Russo AT, et al. An overview of tecovirimat for smallpox treatment and expanded anti-orthopoxvirus applications. Expert Rev Anti Infect Ther. 2021;19(3):331–344.
- Delaune D, Iseni F. Drug development against smallpox: present and future. Antimicrob Agents Chemother. 2020;64:4.
- Hutson CL, et al. Pharmacokinetics and efficacy of a potential smallpox therapeutic, brincidofovir, in a lethal monkeypox virus animal model. mSphere. 2021;6:1.
- Prichard MN, Kern ER. Antiviral activity of 4'-thioIDU and thymidine analogs against orthopoxviruses. Viruses. 2010;2(9):1968–1983.

