ABSTRACT
Hormographiella aspergillata is a rare and emerging cause of invasive mould infections in patients with haematological malignancies, with a mortality rate of approximately 70%. Here, we present the first reported case of suspected disseminated H. aspergillata infection in China. The patient experienced a second relapse of acute myeloid leukaemia and developed neutropenia, fever, discrepant blood pressure between limbs, and cutaneous lesions limited to the left upper extremity. Since lung tissue biopsy was not feasible, metagenomic next-generation sequencing (mNGS) and panfungal polymerase chain reaction (PCR) analysis of bronchoalveolar lavage fluid and blood samples were performed, which indicated probable H. aspergillata pulmonary infection. Histopathology of cutaneous lesions revealed numerous fungal hyphae within dermal blood vessels. mNGS of a skin biopsy sample identified H. aspergillata sequences, and the fungi was subsequently recovered from fungal culture, proving cutaneous H. aspergillata infection. Despite combined antifungal therapy, the patient died owing to disease progression. Additionally, 22 previously reported cases of invasive H. aspergillata infection were reviewed in patients with haematological malignancies. Thus, mNGS is a powerful diagnostic tool for the early and effective detection of invasive H. aspergillata infections, with the advantage of sequencing all potential pathogens, and providing results within 24 h.
Introduction
Although Aspergillus spp. remain the primary cause of invasive mould infections in patients with haematological malignancies, there is an increasing prevalence of non-Aspergillus invasive mould infections in populations with severe and prolonged immunosuppression [Citation1]. Hormographiella aspergillata, the anamorph of Coprinopsis cinerea, is a filamentous basidiomycete commonly found in soil, leaves, press mud compost, and air. Despite its presence in the environment, H. aspergillata rarely causes invasive fungal infections in immunocompromised hosts [Citation2–5]. To date, 22 cases of invasive H. aspergillata infection have been reported in patients with haematological malignancies, with a mortality rate of approximately 70%. The most common clinical manifestations of the disease include pulmonary, rhino-orbito-cerebral, and disseminated infection [Citation6–23].
Invasive H. aspergillata infection diagnosis poses significant challenges in clinical practice. First, the clinical presentation and radiographic manifestations lack specificity. Second, fungal biomarkers including galactomannan and 1,3-β-d-glucan are often negative during invasive H. aspergillata infections [Citation24]. Third, the histopathology of H. aspergillata infection is characterized by the presence of septate hyaline hyphae with a variable width (2–5 μm), branching at an acute angle or dichotomously. However, identifying the fungal genus or species solely based on the histopathological morphology is difficult. [Citation7]. Moreover, culture-based methods have limited sensitivity and may yield negative results despite the presence of fungal elements [Citation22]. Therefore, there is an urgent need for culture-independent assays that can accurately and quickly identify invasive fungi to guide clinical decision-making.
In recent years, significant progress has been made in the development of molecular diagnostic tools to detect invasive H. aspergillata infections. Specifically, panfungal polymerase chain reaction (PCR) followed by sequencing is effective in detecting H. aspergillata in fresh and formalin-fixed paraffin-embedded (FFPE) tissue samples [Citation16,Citation18]. Additionally, metagenomic next-generation sequencing (mNGS), which can characterize all nucleic acids present in a clinical sample, efficiently detects all known/unknown pathogens including bacteria, viruses, and fungi. This is particularly useful for the identification of rare and novel pathogens [Citation25]. However, to date, no reported cases of mNGS identifying infections caused by H. aspergillata have been documented.
Herein, we present a case of suspected disseminated H. aspergillata infection in an immunocompromised patient with lung mass and cutaneous emboli in the left upper extremity. Using mNGS analysis, panfungal PCR, histology, and culture-based methods, we confirmed the suspected disseminated H. aspergillata infection and demonstrated the effectiveness of mNGS for early diagnosis. This is the first reported case of H. aspergillata infection in China to the best of our knowledge; we also reviewed 22 previously reported cases to better understand this rare disease.
Case presentation
A 41-year-old woman was diagnosed with acute myeloid leukemia-M4 and underwent allogeneic hematopoietic stem cell transplantation (HSCT) (). Her first relapse was diagnosed in March 2021, and complete remission was achieved after treatment with donor lymphocyte reinfusion and induction chemotherapy. During consolidation chemotherapy, she developed neutropenia (day 0), which was promptly treated with antifungal prophylaxis using oral posaconazole (200 mg twice daily). Unfortunately, therapeutic drug monitoring (TDM) was unavailable in our clinical laboratory. On day +3, a second relapse was diagnosed, and she underwent a new phase of induction chemotherapy. Considering her persistent neutropenia, she was transferred to an isolation ward to prevent infection (day +11) and a second transplant was planned. Two weeks later, she developed neutropenic fever (day +27), and a chest computerized tomography (CT) scan revealed a mass in the left upper lung. Pulmonary fungal infection was suspected, but blood culture, serum galactomannan and 1,3-β-d-glucan tests were negative. Subsequently, posaconazole prophylaxis was discontinued, and treatment with amphotericin B deoxycholate (AmBd, 25 mg once daily) and broad-spectrum antibiotics were initiated, however, no improvement was observed. Negative blood cultures were also obtained on consecutive days (days +28, 35, 40), while sputum cultures tested positive for Staphylococcus epidermidis (days +35, 40).
Figure 1. Clinical course of the 41-year-old female patient with a disseminated Hormographiella aspergillata infection.
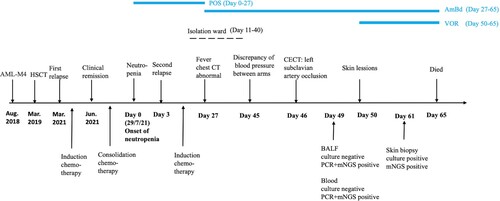
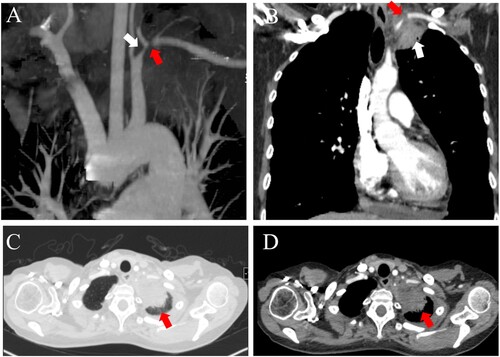
After the patient developed pulmonary infection and bone marrow suppression, the second transplant was postponed, and she was transferred out of the isolation ward and back to the general ward (day +40) to continue anti-infection treatment. Five days later, she began complaining of severe pain in her left upper limb (day +45). Her vital signs showed a blood pressure of 60/30 mm Hg in her left arm and 110/70 mm Hg in her right arm. A bedside ultrasound revealed heterogeneous echogenicity in the left subclavian artery, suggesting a possible embolism in that region. A contrast-enhanced CT scan revealed a mass in the left apex of the lung adjacent to the left subclavian artery (day +46). A low attenuation area was present inside the left subclavian artery, indicating that the mass had extended into the vessel (). On day +49, a bronchoscopy was performed, and the results of fungal culture and galactomannan test of the bronchoalveolar lavage fluid (BALF) sample were negative. However, H. aspergillata sequences were detected in blood and BALF samples using mNGS. Based on these results, intravenous voriconazole (300 mg twice on the first day, followed by 200 mg twice daily) was added to the existing treatment with AmBd.
Figure 2. Contrast-enhanced CT showing a pulmonary mass in the left upper lobe invading the left subclavian artery. CT angiography (A) showing the left subclavian artery stenosis (white arrow) and occlusion (red arrow). CT image of the coronal view (B) showing a mass in the left apex of the lung (white arrow) extending into the left subclavian artery. CT images of the lung window (C) and soft tissue window (D) revealed patchy consolidation in the left apex of the lung (red arrow).
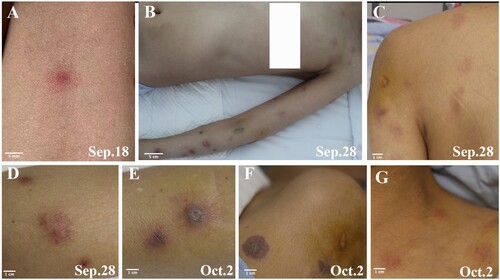
On day +50, an erythematous macule was noticed on the patient's left forearm (), which quickly progressed into painful cutaneous lesions spreading to her left upper extremity. A skin examination revealed a linear distribution of purpuric plaque and subcutaneous nodules on the left arm. Skin biopsy specimens were subjected to mNGS testing, which identified H. aspergillata sequences and the infection was subsequently confirmed by histopathology and fungal culture. Despite maximal medical support, the patient's condition continued to worsen, and she died two weeks after the initial skin lesion appeared, without any neutrophil recovery (day +65). Her family members didn’t provide consent for an autopsy.
Figure 3. Cutaneous lesions evolution throughout disease progression. A: Onset of red papules located on the medial left forearm. B, C: Development of purplish-red plaques and subcutaneous nodules on the left forearm, left anterior chest wall, and left upper back. D: Appearance of erythematous subcutaneous nodules on the inner left forearm. E, F: Presence of a purpuric plaque with necrotic centre on the left forearm. G: Erythematous subcutaneous nodule on the left anterior chest wall.
Materials and methods
Mycology
Blood and BALF samples were subjected to galactomannan and 1,3-β-d-glucan tests. Additionally, blood, sputum, BALF, and skin biopsy samples were streaked onto Sabouraud dextrose agar (SDA) tubes containing chloramphenicol and then incubated at 25 °C for up to three weeks. Positive cultures were identified based on macroscopic and microscopic morphological characteristics, and ITS region sequence analysis. Skin biopsy specimens were stained with haematoxylin and eosin (HE) and Grocott-Gomori methenamine silver without counterstaining.
mNGS analysis
The BALF sample was subjected to PMseq detection at the Beijing Genomics Institute [Citation26]. Briefly, DNA was extracted from 300–500 μL BALF using the TIANamp Micro DNA Kit (Tiangen Biotech, Beijing, China). The extracted DNA was used to produce DNA libraries through DNA fragmentation, end-repair, adapter-ligation, and PCR amplification. Libraries were pooled; DNA Nanoball (DNB) was formed and sequenced by the BGISEQ-50 platform (BGI-Beijing, Beijing, China). Bioinformatics analysis was conducted using the NCBI public database.
Skin-biopsy specimens and blood samples were subjected to mNGS analysis at Hangzhou MatriDx Biotechnology for pathogen detection [Citation27]. DNA extraction and library preparation were performed using the NGS Automatic Library Preparation System (Cat. MAR002, MatriDx Biotech Corp., Hangzhou, China). The libraries were pooled and sequenced on the Illumina NextSeq550 system. Bioinformatic analysis was performed using the NCBI reference sequence database.
Panfungal PCR and sanger sequencing
Genomic DNA was extracted from blood and BALF samples according to the instructions of QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). Amplification was conducted using ITS1 and ITS4 primers targeting the ITS region and NL-1 and NL-4 primers targeting the D1/D2 region [Citation28]. Species identification was performed by sequencing the PCR products and aligning them with DNA sequences in the GenBank database (https://blast.ncbi.nlm.nih.gov/). In cases where two or more PCR products were detected [Citation29], they were cloned and sequenced as previously described [Citation30].
Antifungal susceptibility testing
In vitro antifungal susceptibility testing was performed using CLSI broth microdilution method [Citation31] with some modifications [Citation29]. Antifungals including fluconazole, itraconazole, voriconazole, posaconazole, isavuconazole, caspofungin, anidulafungin, micafungin, and amphotericin B were purchased from Harvey bio (Beijing, China). Isolates were grown on PDA at 37°C for 7 days, and the final inoculum was adjusted to a density of 1× 104–5.0 × 104 hyphal fragments/conidia per mL via counting under a microscope. Plates were incubated at 35°C for 72 h. MIC endpoints were visually determined as the minimum antifungal concentration at which 100% growth inhibition occurred compared to drug-free control wells.
Literature review of invasive H. aspergillata infections in patients with haematological malignancies patients
We conducted a literature search on invasive H. aspergillata infections in patients with haematological cancer, using PubMed (http://www.ncbi.nlm.nih.gov) up to 2022. The search term used was “((Hormographiella aspergillata) OR (Coprinus cinereus)) AND (infection)”. Only invasive H. aspergillata infections in patients with haematological malignancies were included in the analysis. We summarized and analysed epidemiological features, underlying conditions, diagnostic methods, infection sites, treatments, patient prognoses, and antifungal drug sensitivity of the strains. In some cases, no direct biopsy of deep tissue was performed, but we speculated that these sites were infected based on clinical presentation and microbiological examination results and therefore included them in the analysis.
Results
Mycology
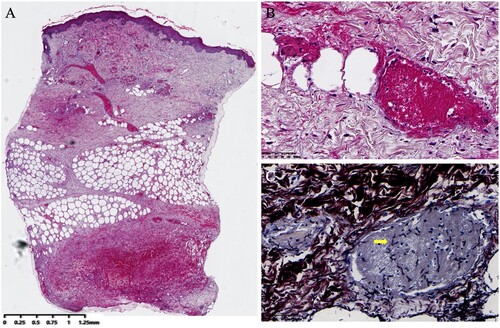
Serial serum (days +28, 35, 42, 49, 62) and BALF samples (day +49) tested negative for galactomannan and 1,3-β-d-glucan. Numerous consecutive blood cultures (days +28, 35, 40, 42, 48, 49, 53, 55) and a BALF cultures (day +49) also tested negative. Two sputum cultures (days +35, 40) tested positive for S. epidermidis. The skin biopsy specimen was cultured on two SDA tubes at 25°C and resulted in the formation of numerous dense, pure, white-coloured colonies with a cotton-like appearance (A). The isolate was further subcultured on an SDA plate at 35°C for 10 days and resulted in white cotton-like colony (B). Microscopic examination of the lactophenol cotton blue mount from the slide culture revealed hyaline, branched, septate hyphae with cylindrical arthroconidia around the conidiophore, but no clamp connections were observed (C). The cultured fungus was further identified via ITS amplification and sequencing; the ITS sequence exhibited 100% identity (650/650 bp) with the corresponding sequence of H. aspergillata CBS 483.71 (GenBank accession no. MH860225.1). The histopathological evaluation of skin lesions revealed thrombosis in dermal and subcutaneous vessels, concomitant with Grocott-Gomori methenamine silver-positive hyphae exhibiting acute angles of branching within vessels ().
Figure 4. Macroscopic and microscopic characteristics of Hormographiella aspergillata. A: Skin tissue cultures on two Sabouraud dextrose agar (SDA) slopes both yielded multiple white, cotton-like, dense pure colonies after seven days of incubation at 25°C. B: White cotton-like colonies on an SDA plate after 10 days of incubation at 35°C. C: Lactophenol cotton blue mount of H. aspergillata slide culture showing branched, septate hyphae with cylindrical arthroconidia accumulating around the conidiophore (original magnification ×400).
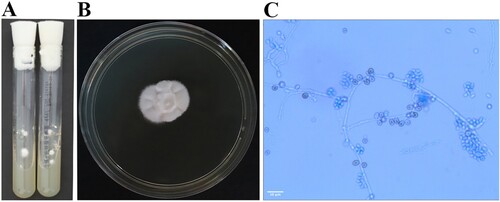
Figure 5. Histological examination of cutaneous Hormographiella aspergillata infection. A: Intravascular thrombosis in both dermal and subcutaneous vessels. (original magnification ×20, haematoxylin and eosin stain). B: Fibrin thrombus in the dermal blood vessels. (original magnification ×400, haematoxylin and eosin stain). C: Branched septate hyphae embedded in fibrin thrombi in the vessels (original magnification ×600, Grocott-Gomori methenamine silver stain).
mNGS analysis
The mNGS analysis of the BALF sample (day +49) detected the presence of H. aspergillata (112 reads) and Aspergillus spp. (60 reads). Later, H. aspergillata (127 reads) but not Aspergillus spp., was detected in the blood sample through mNGS analysis. Subsequently, mNGS analysis of a skin biopsy specimen revealed the presence of H. aspergillata (122,773 reads).
Panfungal PCR and sanger sequencing
The ITS region amplification failed in both blood and BALF samples. When using NL1 and NL4 universal fungal primers, a band of approximately 600 bp was generated in both samples. Direct sequencing of the D1/D2 region of fungi from the blood sample identified the presence of H. aspergillata. Conversely, the Sanger DNA sequencing chromatogram of the PCR product from the BALF sample showed overlapping peaks, making species identification difficult. Therefore, the D1/D2 amplification product was cloned, and 10 positive clones were selected for plasmid DNA extraction and sequencing. This analysis revealed several fungal species in the BALF sample, including Malassezia spp., Cladosporium spp., and H. aspergillata; however, no Aspergillus spp. were detected in either the blood or BALF samples.
Antifungal susceptibility testing
In vitro susceptibility testing results for the isolate are shown in , including the corresponding MICs of fluconazole (64 μg/ml), itraconazole (2 μg/ml), voriconazole (0.5 μg/ml), posaconazole (1 μg/ml), isavuconazole (0.06 μg/ml), caspofungin (>16 μg/ml), micafungin (>16 μg/ml), anidulafungin (4 μg/ml), and amphotericin B (2 μg/ml).
Table 1. Characteristics of invasive Hormographiella aspergillata infection in patients with haematological malignancies.
Literature review of invasive H. aspergillata infections in patients with haematological malignancies patients
Following the literature search, 42 articles were retrieved; 21 articles on H. aspergillata infection were screened, and four articles reporting patients without underlying haematologic disorders were excluded. Thus, total 17 articles were analysed. One case reported in Germany identified the pathogen as Coprinus sp.[Citation7], but not to the species level, and was therefore not retrieved using our search criteria. However, another article identified the strain as similar to H. aspergillata [Citation32]. Consequently, this Germany case [Citation7] was included in the analysis. In total, 22 cases of H. aspergillata infections in patients with haematological cancer described in 18 articles were included in this study, along with the new patient reported herein, resulting in a total of 23 patients ().
Table 2. Antifungal susceptibility profile of Hormographiella aspergillata clinical isolates.
Among the 22 patients, the lungs were the most commonly affected site in 21 cases, followed by the brain (4/22), and the skin and soft tissues (3/22). Furthermore, among these 22 cases, 15 patients had single lung involvement, while seven exhibited disseminated infections involving multiple sites, such as the lungs, skin, brain, eyes, and small intestine.
In total, six cases received antifungal prophylaxis, two patients used posaconazole, while the other four used fluconazole, itraconazole, voriconazole and caspofungin respectively. Although our patient received posaconazole prophylaxis immediately after neutrophil deficiency detection, a breakthrough infection still occurred. Unfortunately, our clinical laboratory lacked TDM, making it impossible to determine whether the patient's plasma posaconazole concentration was below the recommended trough concentration, which could have explained the prophylaxis failure.
The drug sensitivity phenotypes of clinical isolates have been described in several sporadic cases. However, it should be noted that there are no established interpretive breakpoints. In general, voriconazole and amphotericin B exhibit low MICs, while fluconazole and caspofungin exhibit high MICs. Besides, the activities of itraconazole (0.25 to >8 µg/ml), posaconazole (0.06 to >8 µg/ml), isavuconazole (0.06 to >16 µg/ml), anidulafungin (0.12–4 μg/ml), and micafungin (0.25 to >16 μg/ml) are variable.
Discussion
In clinical settings, filamentous basidiomycetes are conventionally classified into Agaricales and Stereales. H. aspergillata, which belongs to the order Agaricales[Citation5,Citation33], has been reported as a cause of invasive fungal disease in patients with haematological malignancies from various regions including Europe, the United States, Colombia, Japan, and Korea. In this study, we report the first case of suspected disseminated H. aspergillata infection in China, to the best of our knowledge.
Pulmonary infection is the most common clinical presentation of invasive H. aspergillata infection (21/22 cases), likely due to the small size of the pathogen's basidiospores that facilitate inhalation and alveolar deposition [Citation5]. Our patient also displayed pulmonary infection with a pulmonary mass on chest CT scan. Furthermore, the patient presented with skin infection. Unlike the previous three case reports with a scattered distribution of skin lesions [Citation10,Citation14,Citation16], this case presented with a linear distribution of skin lesions limited to the left upper extremity, which could be explained by the fungal invasion of the left subclavian artery from adjacent infected tissue in the left apex of the lung, as revealed by ultrasonography and enhanced CT images. This may also account for the discrepant blood pressure between limbs. Notably, a similar case involving Aspergillus flavus was reported by Watsky et al. [Citation34]. In the absence of thrombus and lung tissue biopsies, one could hypothesize that the left subclavian artery thrombus might be a fungal embolus, with the fungus invading the bloodstream and spreading into the distal vessels through the blood, resulting in unilateral cutaneous emboli.
Diagnosing H. aspergillata infection presents significant challenges. A definitive diagnosis requires isolation of the pathogen from a sterile site or histological evidence [Citation35]. Blood culture, which is the gold standard for detecting fungal infections, has low sensitivity, with only one culture positive result among 22 previously reported cases [Citation11]. In our case, eight blood cultures tested consistently negative. Invasive procedures, such as lung tissue biopsy, are rarely feasible for critically-ill patients who cannot tolerate such interventions. In fact, among the 21 reported cases of pulmonary infection, 10 cases did not have lung tissue biopsy including two who underwent BALF analysis, two for whom skin tissue biopsies were performed, and six who were examined postmortem. In our case, skin tissue was obtained for histological and culture analysis, but obtaining the lung tissue was not feasible owing to the patient's frailty and the subsequent refusal of autopsy by family members. Therefore, it is difficult to diagnose H. aspergillata infection by traditional microbiological methods.
The application of molecular techniques for pathogen identification has gained significant importance. Published cases have reported the use of panfungal PCR in diagnosing H. aspergillata in FFEP tissue specimens since 2005 [Citation9], with seven patients being successfully diagnosed using this method. Concurrent amplification of both ITS and D1/D2 regions sometimes may be necessary for detection using panfungal PCR [Citation36]. In this study, successful D1-D2 amplification was achieved, while ITS amplification failed. Chong et al. reported similar results, with pleural fluid specimens demonstrating unsuccessful ITS amplification but successful D1D2 amplification [Citation23]. However, the availability of panfungal PCR testing is limited in China, including many tertiary hospitals [Citation37]. Additionally, in cases of multiple peaks appearing in sequencing results, performing cloning sequencing is necessary, which is a time-consuming and labour-intensive process.
Recently, mNGS has emerged as a valuable diagnostic tool for infectious diseases [Citation38,Citation39]. In this case, mNGS detected a high fungal load of H. aspergillata in the skin biopsy tissue, which was later confirmed by culture and histology, confirming cutaneous H. aspergillata infection. mNGS results of the BALF sample showed the presence of H. aspergillata (112 reads) and Aspergillus spp. (60 reads). However, because BALF is not a sterile sample and both Aspergillus spp. and H. aspergillata are opportunistic pathogens, it is difficult to ascertain whether detected microorganisms are pathogenic, commensal, or environmental contaminants. Negative results of galactomannan and 1,3-β-d-glucan testing suggested that Aspergillus spp. may not be the causative agent. Further mNGS analysis of the blood sample revealed the presence of H. aspergillata (126 reads) without any detection of Aspergillus spp., as confirmed by panfungal PCR. Therefore, the presence of Aspergillus spp. (60 reads) detected in the BALF sample is likely a false positive. Furthermore, the identification of H. aspergillata (112 reads) in BALF and H. aspergillata (126 reads) in blood could potentially be attributed to skin contamination as well. Based on the 2020 EORTC/MSG criteria [Citation35], our patient did not meet the diagnostic criteria for proven invasive pulmonary fungal disease in the absence of lung biopsy and positive blood culture results. However, considering the patient's host factors, abnormal CT-scan findings, and positive panfungal PCR results, a probable diagnosis of invasive pulmonary H. aspergillata infection was made. Furthermore, clinical presentation and serological testing were consistent with the diagnosis, and neither mNGS nor traditional microbiological methods detected any other pathogen related to the patient's condition.
The mNGS technology has significant potential for early detection of invasive H. aspergillata infections. In this case, the patient presented with fever and abnormal pulmonary imaging on day +27 and could not have tolerated lung tissue biopsy due to frailty; blood cultures and serology tests over the following three weeks failed to identify the pathogen responsible for the pulmonary infection. However, on day +49, mNGS successfully identified H. aspergillata, even though blood and BALF cultures tested negative, highlighting the clinical usefulness of mNGS for patients who cannot undergo tissue biopsy. Compared to the traditional fungal culture, which often takes 3–7 days, mNGS provides rapid pathogen detection in <24 h. However, the current cost of mNGS is higher, ¥3000 Renminbi (approximately US$ 500) per sample, compared to fungal culture, which costs 80 Renminbi (approximately US$ 12). Moreover, the lack of standardized criteria for interpreting mNGS results is a considerable challenge and requires collaborations among clinicians, infectious diseases specialists, and clinical microbiologists [Citation40].
A standard treatment for invasive H. aspergillata infection has not yet been established, with a high mortality rate of 72.7% (16/22). In vitro susceptibility and clinical efficacy studies have suggested liposomal amphotericin B (L-AmB) as the first-line antifungal therapy. L-AmB can be administered intravenously with or without inhaled L-AmB, or combined with or without voriconazole, while caspofungin should be avoided [Citation41]. In addition, clinicians should prioritize primary disease treatment and reversing neutropenia, and surgical removal of lesions may potentially improve survival.
In conclusion, we presented the first reported case of suspected invasive H. aspergillata infection affecting the lungs and skin in China. Traditional microbiological methods remain inadequate for detecting H. aspergillata, resulting in delayed diagnoses. Nevertheless, mNGS technology can facilitate early detection of H. aspergillata infections, though further standardization is required.
Ethics statement
This study was performed in compliance with relevant laws and institutional guidelines and was approved by the Ethics Committee of Air Force Medical Center (2021-223-PJ01).
Author contributions
Qian Wang completed molecular experiments, analysed the clinical case, and wrote the manuscript. Xiaoying Yuan designed all the experiments and revised the manuscript. Yinggai Song analysed molecular testing results and revised the manuscript. Qiuhong Yan performed fungal cultures and microscopy tests. Dongmei Han, Hengxiang Wang, Xiaoli Zheng, and Li Ding participated in the treatment of this patient and provided relevant clinical data. Hong Cai and Wei Liu contributed to reviewing the manuscript. All authors have read and approved the final manuscript for publication.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Data supporting the findings of this study are openly available in online repository at: https://www.ncbi.nlm.nih.gov/sra/PRJNA913408.
Correction Statement
This article has been corrected with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Lamoth F, Kontoyiannis DP. Therapeutic challenges of non-Aspergillus invasive mold infections in immunosuppressed patients. Antimicrob Agents Chemother. 2019;63(11):e01244–19. doi:10.1128/AAC.01244-19
- Oliveira TB, Lopes VCP, Barbosa FN, et al. Fungal communities in pressmud composting harbour beneficial and detrimental fungi for human welfare. Microbiology. 2016;162(7):1147–1156. doi:10.1099/mic.0.000306
- Kües U. Life history and developmental processes in the basidiomycete Coprinus cinereus. Microbiol Mol Biol Rev. 2000;64(2):316–353. doi:10.1128/MMBR.64.2.316-353.2000
- Gené J, Guillamón JM, Guarro J, et al. Molecular characterization, relatedness and antifungal susceptibility of the basidiomycetous Hormographiella species and Coprinus cinereus from clinical and environmental sources. Antonie Leeuwenhoek. 1996;70(1):49–57. doi:10.1007/BF00393569
- Chowdhary A, Kathuria S, Agarwal K, et al. Recognizing filamentous basidiomycetes as agents of human disease: a review. Med Mycol. 2014;52(8):782–797. doi:10.1093/mmy/myu047
- Verweij PE, van Kasteren M, van de Nes J, et al. Fatal pulmonary infection caused by the basidiomycete Hormographiella aspergillata. J Clin Microbiol. 1997;35(10):2675–2678. doi:10.1128/jcm.35.10.2675-2678.1997
- Nenoff P, Friedrich T, Schwenke H, et al. Rare fatal simultaneous mould infection of the lung caused by Aspergillus flavus and the basidiomycete Coprinus sp. in a leukemic patient. J Med Vet Mycol. 1997;35(1):65–69. doi:10.1080/02681219780000901
- Surmont I, Van Aelst F, Verbanck J, et al. A pulmonary infection caused by Coprinus cinereus (Hormographiella aspergillata) diagnosed after a neutropenic episode. Med Mycol. 2002;40(2):217–219. doi:10.1080/mmy.40.2.217.219
- Lagrou K, Massonet C, Theunissen K, et al. Fatal pulmonary infection in a leukaemic patient caused by Hormographiella aspergillata. J Med Microbiol. 2005;54(7):685–688. doi:10.1099/jmm.0.46016-0
- Abuali MM, Posada R, Del Toro G, et al. Rhizomucor variabilis var. regularior and Hormographiella aspergillata infections in a leukemic bone marrow transplant recipient with refractory neutropenia. J Clin Microbiol. 2009;47(12):4176–4179. doi:10.1128/JCM.00305-09
- Conen A, Weisser M, Hohler D, et al. Hormographiella aspergillata: an emerging mould in acute leukaemia patients? Clin Microbiol Infect. 2011;17(2):273–277. doi:10.1111/j.1469-0691.2010.03266.x
- Suarez F, Olivier G, Garcia-Hermoso D, et al. Breakthrough Hormographiella aspergillata infections arising in neutropenic patients treated empirically with caspofungin. J Clin Microbiol. 2011;49(1):461–465. doi:10.1128/JCM.01213-10
- Pang KA, Godet C, Fekkar A, et al. Breakthrough invasive mould infections in patients treated with caspofungin. J Infect. 2012;64(4):424–429. doi:10.1016/j.jinf.2011.12.015
- Bojic M, Willinger B, Rath T, et al. Fatal skin and pulmonary infection caused by Hormographiella aspergillata in a leukaemic patient: case report and literature overview. Mycoses. 2013;56(6):687–689. doi:10.1111/myc.12087
- Corzo-León DE, Satlin MJ, Soave R, et al. Epidemiology and outcomes of invasive fungal infections in allogeneic haematopoietic stem cell transplant recipients in the era of antifungal prophylaxis: a single-centre study with focus on emerging pathogens. Mycoses. 2015;58(6):325–336. doi:10.1111/myc.12318
- Heiblig M, Bozzoli V, Saison J, et al. Combined medico-surgical strategy for invasive sino-orbito-cerebral breakthrough fungal infection with Hormographiella aspergillata in an acute leukaemia patient. Mycoses. 2015;58(5):308–312. doi:10.1111/myc.12305
- Nanno S, Nakane T, Okamura H, et al. Disseminated Hormographiella aspergillata infection with involvement of the lung, brain, and small intestine following allogeneic hematopoietic stem cell transplantation: case report and literature review. Transpl Infect Dis. 2016;18(4):611–616. doi:10.1111/tid.12561
- Godet C, Cateau E, Rammaert B, et al. Nebulized liposomal amphotericin B for treatment of pulmonary infection caused by Hormographiella aspergillata: case report and literature review. Mycopathologia. 2017;182(7-8):709–713. doi:10.1007/s11046-017-0117-9
- Chauhan A, Gruenberg J, Arbefeville S, et al. Disseminated Hormographiella aspergillata infection with lung and brain involvement after allogenic hematopoietic stem-cell transplantation in a 54-year-old man. Lab Med. 2019;50(4):426–431. doi:10.1093/labmed/lmz018
- Isabel Cristina RS, Diana A, Karen A. Breakthrough Hormographiella aspergillata infection in a patient with acute myeloid leukemia receiving posaconazole prophylaxis: A case report and review. Mycopathologia. 2020;185(6):1069–1076. doi:10.1007/s11046-020-00488-z
- Moniot M, Lavergne RA, Morel T, et al. Hormographiella aspergillata: an emerging basidiomycete in the clinical setting? A case report and literature review. BMC Infect Dis. 2020;20(1):945. doi:10.1186/s12879-020-05679-z
- Tschopp J, Perentes JY, Beigelman-Aubry C, et al. Invasive Hormographiella aspergillata infection in patients with acute myeloid leukemia: report of two cases successfully treated and review of the literature. Med Mycol Case Rep. 2021;32:68–72. doi:10.1016/j.mmcr.2021.03.008
- Chong E, Yu HJ, Kim TY, et al. Invasive hormographiella aspergillata infection identified using DNA sequencing. Ann Lab Med. 2022;42(3):370–372. doi:10.3343/alm.2022.42.3.370
- Delliere S, Rivero-Menendez O, Gautier C, et al. Emerging mould infections: get prepared to meet unexpected fungi in your patient. Med Mycol. 2020;58(2):156–162.
- Han D, Li Z, Li R, et al. mNGS in clinical microbiology laboratories: on the road to maturity. Crit Rev Microbiol. 2019;45(5-6):668–685. doi:10.1080/1040841X.2019.1681933
- Zhou X, Wu H, Ruan Q, et al. Clinical evaluation of diagnosis efficacy of active mycobacterium tuberculosis complex infection via metagenomic next-generation sequencing of direct clinical samples. Front Cell Infect Microbiol. 2019;9:351. doi:10.3389/fcimb.2019.00351
- Guo Y, Li H, Chen H, et al. Metagenomic next-generation sequencing to identify pathogens and cancer in lung biopsy tissue. EBiomedicine. 2021;73:103639. doi:10.1016/j.ebiom.2021.103639
- Chowdhary A, Agarwal K, Kathuria S, et al. Clinical significance of filamentous basidiomycetes illustrated by isolates of the novel opportunist Ceriporia lacerata from the human respiratory tract. J Clin Microbiol. 2013;51(2):585–590. doi:10.1128/JCM.02943-12
- Trubiano JA, Dennison AM, Morrissey CO, et al. Clinical utility of panfungal polymerase chain reaction for the diagnosis of invasive fungal disease: a single center experience. Med Mycol. 2016;54(2):138–146. doi:10.1093/mmy/myv092
- Yang X, Song Y, Liang T, et al. Application of laser capture microdissection and PCR sequencing in the diagnosis of Coccidioides spp. infection: A case report and literature review in China. Emerg Microbes Infect. 2021;10(1):331–341. doi:10.1080/22221751.2021.1889931
- Clinical and Laboratory Standards Institute (CLSI). Reference method for broth dilution antifungal susceptibility testing of filamentous fungi. 3rd ed. CLSI standard M38 (ISBN 1-56238-830-4 [Print]; ISBN 1-56238-831-2 [Electronic]). Clinical and Laboratory Standards Institute, Wayne, PA, 2017.
- De Hoog GS, Gerrits van den Ende AH. Molecular diagnostics of clinical strains of filamentous basidiomycetes. Mycoses. 1998;41(5–6):183–189. doi:10.1111/j.1439-0507.1998.tb00321.x
- Chowdhary A, Agarwal K, Meis JF. Filamentous fungi in respiratory infections. What lies beyond Aspergillosis and Mucormycosis? PLOS Pathog. 2016;12(4):e1005491. doi:10.1371/journal.ppat.1005491
- Watsky KL, Eisen RN, Bolognia JL. Unilateral cutaneous emboli of Aspergillus. Arch Dermatol. 1990;126(9):1214–1217. doi:10.1001/archderm.1990.01670330094015
- Donnelly JP, Chen SC, Kauffman CA, et al. Revision and update of the consensus definitions of invasive fungal disease from the European organization for research and treatment of cancer and the mycoses study group education and research consortium. Clin Infect Dis. 2020;71(6):1367–1376. doi:10.1093/cid/ciz1008
- Lau A, Chen S, Sorrell T, et al. Development and clinical application of a panfungal PCR assay to detect and identify fungal DNA in tissue specimens. J Clin Microbiol. 2007;45(2):380–385. doi:10.1128/JCM.01862-06
- Li N, Li S, Tan W, et al. Metagenomic next-generation sequencing in the family outbreak of psittacosis: the first reported family outbreak of psittacosis in China under COVID-19. Emerg Microbes Infect. 2021;10(1):1418–1428. doi:10.1080/22221751.2021.1948358
- Wilson MR, Naccache SN, Samayoa E, et al. Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N Engl J Med. 2014;370(25):2408–2417. doi:10.1056/NEJMoa1401268
- Chiu CY, Miller SA. Clinical metagenomics. Nat Rev Genet. 2019;20(6):341–355. doi:10.1038/s41576-019-0113-7
- Stratton CW, Schutzbank TE, Tang YW. Use of metagenomic next-generation sequencing in the clinical microbiology laboratory: A step forward, but not an end-all. J Mol Diagn. 2021;23(11):1415–1421. doi:10.1016/j.jmoldx.2021.09.003
- Hoenigl M, Salmanton-García J, Walsh TJ, et al. Global guideline for the diagnosis and management of rare mould infections: an initiative of the European confederation of medical mycology in cooperation with the international society for human and animal mycology and the American society for microbiology. Lancet Infect Dis. 2021;21(8):e246–e257. doi:10.1016/S1473-3099(20)30784-2

