ABSTRACT
Macroautophagy (also known as autophagy) plays a pivotal role in maintaining cellular homeostasis. The terminal step of the multi-step autophagy degradation pathway involves fusion between the cargo-laden, double-membraned autophagosome and the lytic organelle lysosome/vacuole. Over the past decade, various core components of the molecular machinery that execute this critical terminal autophagy event have been identified. This review highlights recent advances in understanding the molecular structures, biochemical functions, and regulatory mechanisms of key components of this highly sophisticated machinery including the SNARE fusogens, tethering factors, Rab GTPases and associated guanine nucleotide exchange factors, and other accessory factors.
Introduction
Macroautophagy (hereafter referred to as autophagy) plays a key role in the maintenance of cellular homeostasis by removing protein aggregates, generating nutrients during stress, and turning over damaged organelles. The multi-step autophagic degradation process begins with the formation of a crescent-shaped membrane structure known as the phagophore. The phagophore wraps around a cargo and self-fuses into a double-membrane transport vesicle called the autophagosome. The cargo-laden autophagosome ultimately fuses with the lysosome or vacuole where the content is degraded inside the lumen by lysosomal enzymes and the macromolecules are recycled by lysosomal permeases. The discovery of the ATG (autophagy-related) genes by yeast genetic screens in the 1990s propelled molecular investigations of the autophagy pathway. Subsequent biochemical, structural, and cell biology-based studies on the core ATG proteins have generated insights into the molecular mechanism of autophagy initiation and autophagosome biogenesis in different model organisms and in mammalian cells. In recent years, there has been significant progress made in understanding the molecular details of late autophagy events. The most notable findings are the identification of specific Rab (Ras-associated binding) GTPases, guanine nucleotide exchange factors (GEF), SNARE (Soluble N-ethylmaleimide-sensitive factor attachment protein receptor) proteins, and tethering factors that function collaboratively in a similar fashion as their counterparts in conventional intracellular trafficking pathways to mediate fusion between the autophagosome and lysosome/vacuole (). Several excellent recent reviews have covered major findings in this area of autophagy research.Citation1–5 This review is dedicated to describing advances made in understanding the biochemical features and molecular structures of components of the autophagic fusion machinery.
Table 1. Components of the autophagosome-lysosome/vacuole fusion machinery.
SNARE proteins
Similar to other membrane trafficking processes, SNARE proteins represent the main driver of autophagome-lysosome/vacuole fusion in autophagy.Citation6 In particular, various SNARE proteins localized to opposing membranes, and when brought within a distance of 10nm, are capable of “zippering” themselves into a four-helical bundle that can bring two membranes closer and reduce the energy barrier for membrane fusion.Citation7,Citation8 Based on the presence of a conserved glutamate (Q) or arginine (R) residue in the highly conserved “zero layer”, SNARE proteins can be classified into three Q-SNARE proteins – Qa, Qb, and Qc – and one R-SNARE protein.Citation9 Despite considerable sequence divergence, their mechanism of action is conserved; different sets of SNARE proteins assemble into complexes (i.e., QabcR-complex) in a combinatorial fashion that has been suggested to contribute to fusion specificity.Citation10,Citation11 All SNARE complexes contain tail-anchored members, which have cytosolic N-terminal fragments and a single transmembrane domain (TMD) or double TMDs at the end of the C-terminus.Citation12,Citation13 Some SNARE proteins are cytosolic and collaborate with membrane-anchored members for the formation of SNARE protein complexes.Citation14
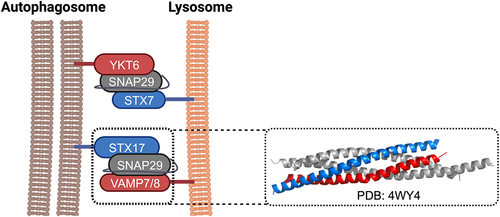
It has been suggested that Vam3, Vam7, Ykt6, and Vti1 are involved in autophagosome-vacuole fusion in yeast and that vesicle-associated membrane protein 7 (VAMP7), VAMP8, and Vti1 are responsible for autophagosome-endolysosome fusion in mammals ().Citation15–17 Meanwhile, a set of SNARE proteins, including syntaxin 17 (STX17), synaptosomal-associated protein 29 (SNAP29) on autophagosomes, and VAMP8 on lysosomes, have been found to form SNARE complexes that are essential for the fusion between autophagosomes and lysosomes.Citation18,Citation19 Crystallographic analysis revealed that the SNARE domains of STX17, SNAP29, and VAMP8 form a helical bundle that resembles other previously characterized trans-SNARE complexesCitation20 (). The Q-SNARE protein STX17 translocates to the outer membrane of the completed autophagosome, and the packed hairpin-like structure causes the exposure of hydrophobic residues.Citation18 In contrast to other SNARE proteins, the unique hairpin-like double TMDs localize STX17 to autophagosomes and reduce fusion rate by increasing the protein-membrane mismatch.Citation21 Besides STX17 and SNAP29 on autophagosomes and VAMP8 on endolysosomes, recently, another independent set of SNAREs, YKT6/SNAP29 on autophagosomes and syntaxin 7 (STX7) on endolysosomes, has also been proposed to mediate lysosome–autophagosome fusion.Citation22 Meanwhile, for the late stage autophagosome–vacuole fusion, the R-SNARE Ykt6 on the autophagosome and the Q-SNAREs Vam3, Vam7, and Vti1 on the vacuole have been recently found to play an important role in yeastCitation23 and in Drosophila melanogaster.Citation24
Figure 1. Mammalian autophagic SNARE proteins. Two sets of SNAREs capable of forming trans-SNARE complexes have been implicated in autophagosome-lysosome fusion in mammalian cells: (1) syntaxin 17 (STX17)/ SNAP29/ VAMP7(8) and (2) Ykt6/ SNAP29/ syntaxin 7 (STX7). The Qa, Qbc, and R SNAREs are coloured in blue, grey, and red, respectively. The crystal structure of the complex formed by the SNARE domains of STX17, SNAP29, and VAMP8 is displayed on the right.
Post-translational modifications (PTMs), such as phosphorylation, of SNAREs have been shown to regulate membrane fusion by impacting SNARE zipping.Citation25 Particularly, PTMs at the residues of SNARE domain facing towards ionic layers can block SNARE zipping.Citation26 O-linked β-N-acetylglucosamine (O-GlcNAc) transferase (OGT) mediates the O-GlcNAcylation of SNAP29 and regulates autophagy in a nutrient-dependent manner. In mammalian cells, OGT knockdown, or the mutation of O-GlcNAc sites in SNAP29, promotes the formation of a SNAP29-containing a SNARE protein complex, increases fusion between autophagosomes and both endosomes and lysosomes, and promotes autophagic flux.Citation27 It has been reported that O-GlcNAc-modified SNAP29 reduces the binding affinity with partner SNARE proteins and thus attenuates the assembly of the SNAP29-containing SNARE protein complex. O-GlcNAc-modification of SNAP29 could create steric hindrance that affects SNARE assembly and function, thus preventing the untimely or ectopic formation of the SNARE protein complex. Remarkably, the depletion of ogt-1 has a similar effect on autophagy in Caenorhabditis elegans, while the expression of an O-GlcNAc-defective SNAP29 mutant facilitates the autophagic degradation of protein aggregates.Citation28 Most recently, the fusion blocked by the O-GlcNAcylation of SNAP29 has been shown to promote apoptosis via ROS production.Citation29 The phosphorylation on VAMP8 has been shown to influence membrane fusion.Citation25 It was recently shown that VAMP8 plays an important role in forming a prefusion state of lysosomal clusters, in which multiple lysosomes form clusters around individual autophagosomes, setting the stage for membrane fusion.Citation30 Using a phosphorylation mimic for C-terminal residues of VAMP8, researchers observed a decrease of fusion in an ensemble lipid mixing assay and an increase of unfused lysosomes associated with autophagosomes.Citation30 These results suggest that phosphorylation not only reduces spontaneous fusion for minimizing autophagic flux under normal conditions, but also preassembles multiple lysosomes to increase the fusion probability for resuming autophagy upon stimulation. A parallel study identified mTOR as the kinase for VAMP8 phosphorylation.Citation31 STX17 contains sites for both acetylationCitation32 and phosphorylation.Citation33 Two residues, K219 and K223, on STX17 are acetylated by CREB binding protein under normal conditions to block SNARE zippering and thereby minimize autophagy flux. Similar to the VAMP8 dephosphorylation, upon stimulation, the deacetylation of STX17 by HDAC2 can release the brake of SNARE zipping and significantly increase autophagy.Citation32 The phosphorylation of STX17 at the residue S200 by TBK1 is mainly for regulating the formation of the autophagy initiation ATG13 complex.Citation33 Finally, ULK1 keeps the inactivated YKT6 through phosphorylation until the completion of autophagosome formation,Citation34–36 which prevents the formation of premature complexes.
Tethering factors
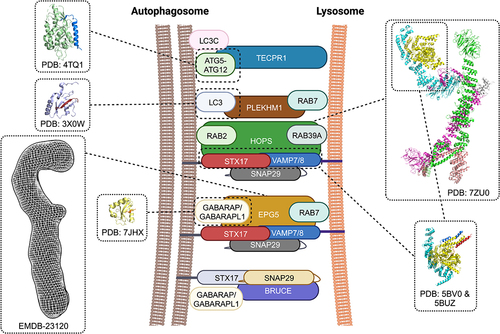
SNARE proteins alone are not sufficient to drive membrane fusion and require other accessary factors to facilitate this process. Perhaps the most important accessory factors are tethering factors, which are recruited to specific membranes by Rab GTPases and phosphoinositide lipids to mediate initial engagement between an incoming vesicle and its target organelle.Citation37,Citation38 The action of tethering factors is thought to bring the two compartments to a sufficiently close distance to allow trans-SNARE assembly. It has also been postulated that tethering factors confer fusion efficiency and specificity by binding/chaperoning distinct SNARE proteins and/or SNARE subassemblies to promote trans-SNARE formation.Citation39 Studies in conventional membrane trafficking pathways have identified two major groups of tethering factors: long coiled-coil tethers and multi-protein tethering complexes (MTC’s).Citation38 Amongst these, the HOPS (homotypic fusion and protein sorting) complex, an MTC that mediates endosomal trafficking,Citation40 has been demonstrated to function in autophagosome-lysosome tethering.Citation41,Citation42 Recent investigations in mammalian cells have identified several other noncanonical tethering factors that are critical to autophagosome-lysosome fusion (, ).
Figure 2. Accessory factors that function in autophagosome-lysosome fusion. TECPR1 binds LC3C to promote fusion between autophagosomes and lysosomes. The crystal structure of the human TECPR1 AIR domain in complex with human ATG5 (TECPR1 AIR domain in blue, ATG5 in light green) showed that TECPR1 binds to the same interface that ATG5 uses to bind ATG16L. The Rab7 effector PLEKHM1 functions as a multivalent adaptor to mediate autophagosome-lysosome fusion. Human PLEKHM1 binds LC3 proteins through a conserved LIR domain and this interaction has been visualized by X-ray crystallography (LC3 in light blue, PLEKHM1 LIR motif in brown). The highly conserved HOPS complex facilitates autophagosome-lysosome fusion by tethering membranes, chaperoning and proofreading SNAREs, and promoting trans-SNARE assembly. The overall architecture of yeast Saccharomyces cerevisiae HOPS was visualized by cryo-EM and an atomic model of the core complex was generated based on the cryo-EM density map (Vps33a in yellow, Vps16 in cyan, Vps11 in green, Vps8 in magenta, Vps39 in salmon, Vps41 in grey). Two crystal structures of Chaetomium thermophilum Vps33-Vps16 subassembly in complex with the Qa-SNARE Vam3 and the R-SNARE Nyv1 revealed how the SM-module of HOPS bind SNAREs. A composite model generated from these two crystal structures is shown in the inset with Vam3 coloured in blue and Nyv1 coloured in red. EPG5 is thought to tether autophagosomes to lysosomes through binding LC3/GABARAP proteins and Rab7. The overall architecture of full-length human EPG5 has been visualized by negative stain electron microscopy (density map coloured in grey). X-ray crystallography revealed how the LIR2 motif of human EPG5 (coloured in red) interacts with GABARAPL1 (coloured in yellow).
The HOPS complex
Conserved from yeast to humans, HOPS mediates the fusion of late endosomes and autophagosomes with lysosomes/vacuoles by tethering membranes, chaperoning, proofreading SNARE’s and promoting trans-SNARE assembly.Citation43 Yeast HOPS binds the Q-SNAREs Vam3, Vam7, and Vti1 and R-SNARE Nyv1Citation44 while mammalian HOPS binds the autophagosomal Qa-SNARE STX17Citation41,Citation42 with conflicting data regarding its ability to bind the autophagosomal R-SNARE Ykt6.Citation22,Citation23 HOPS is thought to be recruited to autophagosomes, late endosomes, and lysosomes/vacuoles directly by Ypt7 in yeast or indirectly by the small GTPase RAB7 through the adaptor protein PLEKHMI in higher eukaryotes.Citation45 Phosphoinositide lipids might also play a role in HOPS recruitment as this complex shows an affinity for PI(3)P, PI(3,5)P2, and PI(4,5)P2 in vitro.Citation46 In mammalian cells, HOPS has also been shown to interact with the GABARAP subfamily of Atg8 autophagosome surface proteins and to bind lysosomes via the ARL8B small GTPase and the BORC complex.Citation45,Citation47,Citation48
The hexameric HOPS complex is composed of two HOPS-specific subunits (Vps39 and Vps41) and four core subunits (Vps11, Vps18, Vps16, and Vps33) shared with the related CORVET complex that functions in early endosomal trafficking.Citation43 X-ray crystallography generated the first structural information on HOPS, and in particular, the Vps33 subunit that serves as a SM (Sec1/Munc18) protein that chaperones SNARE proteins and facilitates trans-SNARE assembly. The first crystal structures of human Vps33a and Vps33 from the thermophilic fungus Chaetomium thermophilium not only showed that this HOPS subunit adopts an overall fold similar to other structurally characterized SM proteins but also revealed that this SM protein does not contain a binding groove for the N-terminal peptide of Qa-SNARE.Citation49 The subsequent crystal structures of Vps33 proteins in complex with the C-terminal domain (CTD) of the HOPS subunit, which is the first structural data obtained for a HOPS subassembly, demonstrated that Vps33 binds the alpha-solenoid of the Vps16 CTD via an extended surface.Citation50 Lastly, a landmark crystallographic study generated structural snapshots of Vps33 in complex with the SNARE domains of the R-SNARE Nyv1 or the Qa-SNARE Vam3. These structures revealed that the SNARE motifs of Nyv1 and Vam3 adopt helical structures and bind to overlapping sites. When the two structures were superimposed, the two SNARE domains are arranged in a half-zippered configuration with their zero-layer residues located proximal to one another.Citation51 This finding led to the proposal that Vps33 not only templates the folding of the two complementary SNARE domains from the Qa- and R-SNAREs but also guides them toward complex assembly (). Future studies will focus on delineating how Vps33 assists the observed assembly intermediate to incorporate the third SNARE and to transition into the full assembly, and to determine if mammalian VPS33a exerts similar SNARE chaperoning/templating function on its cognate autophagic SNARE’s STX17 and VAMP7/8.
The other five HOPS subunits share a similar overall architecture featuring N-terminal beta-propeller domains followed by extended alpha-solenoids similar to those found in clathrin, COP-II cage components, and coat nucleoporins. Except for the N-terminal beta-propeller of yeast Vps18Citation52, experimental structural information has been difficult to obtain for HOPS subunits through the X-ray crystallographic approach. The development of procedures to isolate native and overexpressed HOPS from yeast cells has facilitated structural studies of this MTC in its fully assembled state by the single-particle electron microscopy (EM) approach. The first negative stain EM investigation of full yeast HOPS, which involves stabilization by mild chemical crosslinking, revealed a seahorse-like architecture featuring a head region containing a deep cavity and attached to a tail region that contains a flexible lobe.Citation53 Further negative stain EM analysis of HOPS subassemblies enabled these investigators to deduce a subunit organization map of the complex, with the Vps33, Vps16, and Vps41 making up the head domain, Vps11 and Vps18 making up the extended tail, and Vps39 the tail-localized flexible lobe. They also demonstrated that the Vps41 and Vps39 subunits located at the tips of the complex can engage in interaction with Ypt7. Interestingly, a subsequent negative stain EM study on non-crosslinked yeast HOPS reported by a different research group showed that this complex adopts a more extended “spaghetti-dancer” architecture featuring a “head region” composed of two domains and three leg-like extensions.Citation54 As the proposed subunit organization of the two EM studies largely agrees with one another, one suggestion is that the two studies captured yeast HOPS in two different conformational states: open and closed.
The recent breakthrough single-particle cryo-EM investigation reported, for the first time, visualization of the complete yeast HOPS complex in its solution state (). Although they only managed to obtain a medium-resolution (local resolution between 3.6 and 5 Å) composite map by combining several local-refined maps, these investigators were able to construct an atomic model of yeast HOPS by leveraging the power of the recently public-released machine learning-based AlphaFold2 structural prediction algorithm.Citation55 In particular, these investigators used AlphaFold2 to generate structural models of all HOPS subunits and then fit them into their cryo-EM density map. The refined structural model showed that yeast HOPS adopts a highly extended yet relatively rigid architecture of 430 Å in height and 130 Å in width, with an overall morphology resembling a baseball pitcher. The two core subunits Vps11 and Vps18 engage in interactions with one another through their elongated alpha-solenoids in an antiparallel fashion to form the rigid “body” or central core, while their N-terminal beta-propeller domains project to the periphery forming the “head” (Vps11) of one of the “feet” on the opposite side of the complex. On the other hand, the HOPS-specific units Vps39 and Vps41 are anchored to the core by their C-terminal helical regions through engaging in coiled-coil interactions with the extended C-terminal helix and the RING finger domain of Vps11 and Vps18 respectively. Even though the rest of Vps39 and Vps41 are poorly resolved in the cryo-EM map beyond the regions in contact with the core due to their inherent flexibility, the AlphaFold2-predicted structures suggested that Vps39 forms the second leg and feet while Vps41 forms one of the arms and hands. Lastly, the SNARE binding module subunits Vps16 and Vps33 form the second arm and hand holding the “baseball”. More specifically, Vps16 extends to the lateral side of the core and engages in an interaction interface through part of its alpha-solenoid with a coiled coil formed by Vps18 and Vps41 and the N-terminal region of Vps18. The SM protein Vps33, however, is anchored to the alpha-solenoid of Vps16 and also contacts a structured loop of Vps18. These investigators also used AlphaFold2 to model the interaction between Vps39 and Vps41 and Ypt7 GTPase and found that Ypt7 binds the beta-propeller of Vps41 and the alpha-solenoid of Vps39, respectively. Based on their new structural findings, these investigators proposed a model in which the Vps41 and Vps39 recruit HOPS to opposing membranes to allow the rigid central core of HOPS to stably tether these membranes. In the next phase, Vps33 engages in interaction with the SNAREs to promote zippering with the slight flexibility of Vps41 and Vps39 allowing them to dampen the motion of HOPS as the opposing membranes are brought closer together.
Whether or not mammalian HOPS adopts a similar architecture remains unknown. However, it is worth mentioning that a very recent study proposed a “hook-up” model for mammalian HOPS complex assembly, which requires two HOPS sub-complexes docking on membranes via membrane-associated Rab GTPases.Citation56 The four-subunit subcomplex containing VPS16/VPS18/VPS33A/VPS41 binds with Rab39A on lysosomes via its VPS41, while the two-subunit HOPS subcomplex containing VPS39/VPS11 binds with Rab2 on autophagosomes via its subunit VPS39. Proper pairing with Rab2 and Rab39A enables HOPS complex assembly between autophagosome and lysosome for its tethering function, facilitating efficient membrane fusion driven by autophagic SNAREs.
EPG5
Originally discovered in the C. elegans genetic screen metazoan-specific autophagy factors,Citation57 EPG5 (Ectopic P-granules protein 5) is a Rab7 effector that functions as a tethering factor in autophagosome fusion with late endosome/lysosome.Citation58 Deficiency in EPG5 in C. elegans, mice, and humans leads to the accumulation of autophagosomes, amphisomes, and non-degradative autolysosomes and in turn impairment in autophagy.Citation58 Recombinant C. elegans EPG-5 and human hEPG5 promote assembly of the STX17-SNAP29-VAMP8 trans-SNARE complex in vitro and knockdown of EPG5 in HeLa cells reduces levels of STX17, SNAP29, VAMP7, and VAMP858. EPG5 engages with autophagosomes through interaction with LC3/GABARAP family of ATG8 proteins via its two tandemly arranged LIR motifs, and is likely localized to the late/lysosomes through Rab7 and/or the R-SNARE VAMP7/8. EPG-5 deficiency leads to non-specific fusion of autophagosomes with different endocytic vesicles and the formation of abnormally large vesicles with mixed identities in C. elegans, indicating that this tethering factor plays a pivotal role in enforcing fusion specificity.Citation58 Mutations to human EPG5 cause an autosomal recessive severe multi-system disorder known as Vici syndrome.Citation59
Recent studies have generated new insights into the biochemical and structural properties of human EPG5. Negative stain EM analysis revealed that the ~290kDa human EPG5 adopts an overall shepherd staff architecture with a “hook” connected to a “finger region” and rigid shaftCitation60 (). Its highly extended nature and overall length (375Å) are in line with that observed for MTCs in other membrane trafficking pathways. Biochemical pulldown analysis showed that EPG5 preferentially binds the GABARAP subfamily of ATG8 proteins. Although both LIR motifs within the tandem are required for maximal binding to GABARAPs, one LIR (LIR2) exhibits stronger binding affinity compared to the other (LIR1). The crystal structure of EPG5 LIR2 in complex with GABARAPL1 revealed that this LIR docks to the canonical LDS (LIR docking site) on this human ATG8 isoformCitation60 (). Further negative stain EM studies suggested that this tandem LIR is localized to the concave side of the hook region of EPG5. Finally, mutagenesis studies showed that despite being the low-affinity binding site, LIR1 plays a more important role in mediating interaction with GABARAP. Collectively, these data led to a two-factor authentication model in which the initial binding of LIR1 to GABARAP licensed the subsequent docking and binding of LIR2 to lock EPG5 on autophagosomes into a tight interaction in preparation for subsequent fusion with the lysosome.Citation6
Other accessory factors
ATG14/Barkor/ATG14L, an essential autophagy-specific regulator of the class III phosphatidylinositol 3-kinase (PtdIns3K) complex, is concentrated on the curved autophagic membrane enriched in PtdIns(3)P via the Barkor/ATG14(L) autophagosome targeting sequence (BATS) domain in a stress-inducible manner.Citation61,Citation62 That finding indicates that Barkor/ATG14(L) functions as a membrane curvature sensor and targeting factor for PI3KC3 to autophagosomes recruited by STX17.Citation61 In addition to its localization to phagophores,Citation62–66 ATG14 also localizes to mature autophagosomes and controls the fusogenic activity of the autophagic SNARE protein complex, both spatially and temporally.Citation20,Citation61,Citation63,Citation67 It has been previously shown that human ATG14 binds directly to the binary complex of STX17 and SNAP29 on autophagosomes and promotes full SNARE complex zippering to mediate the fusion between autophagosomes and lysosomes. ATG14 homo-oligomerization is required for SNARE protein binding and fusion promoting, yet is dispensable for PtdIns3K stimulation and autophagosome biogenesis.Citation20 These data suggest ATG14’s pivotal role in autophagy-specific autophagosome–endolysosome fusion activity.
TECPR1 was initially identified component of the autophagy network via its interaction with Atg5 from proteomics-based interactome analysis.Citation68 TECPR1 co-localized with lysosomal LAMP-2 and deficiency in this protein leads to accumulation of autophagosomes. Subsequent analysis revealed that TECPR1 interacts with the Atg12-Atg5 conjugate via the AIR (Atg12-Atg5-interacting region) domain located adjacent to a PH (pleckstrin homology) domain that mediates PI3P binding. TECPR1 binds LC3C to promote fusion between autophagosomes and lysosomes. The crystal structure of human TECPR1 AIR domain in complex with human ATG5 revealed that TECPR1 binds to the same interface as ATG16L which forms a complex with Atg12-Atg5 conjugateCitation69 (). This leads to a working model in which Atg16L hands off the Atg12-Atg5 conjugate to TECPR1 and this interaction leads to a conformational change that exposes the PH domain of TECPR1 for binding PI3P on autophagosome to confer fusion specificity. Interestingly, three different research groups (Randow, Wu, Lystad) recently discovered that TECPR1 contains an N-terminal dysferlin domain that binds sphingomyelin and likely serves as an E3 ligase to mediate conjugation of ATG8 proteins to single membranes induced by membrane damage.Citation70–73
A member of the inhibitor of apoptosis (IAP) family, BRUCE is a large-sized protein that was identified by an RNAi screen for autophagy factors. BRUCE localizes to both lysosomes and autophagosomes. Deficiency in BRUCE leads to the accumulation of autophagosomes upon starvation and in turn defective autophagy.Citation74 BRUCE binds the autophagic snares STX17 and SNAP29 and it engages GABARAP and GABARAPL1 in a LIR-independent manner.Citation74 Future studies will focus on mapping the GABARAP-binding site on BRUCE and on delineating how it engages in non-canonical interaction with Atg8 proteins.
PLEKHM1 (pleckstrin homology domain containing, family M [with RUN domain] member 1) is a Rab7 effector discovered from a proteomics analysis of interaction partners of active Rab7.Citation71 It serves as a multivalent adaptor to regulate fusion between autophagosome and lysosome by simultaneously binding Rab7, Arl8, LC3, and the HOPS complex.Citation75 PLEKHM1 binds LC3/GABARAP proteins through a conserved LIR domain. Crystallographic analysis revealed that PLEKHM1’s LIR docks into the LDS of LC3B in a canonical fashionCitation45 (). PLEKHMI has also been shown to interact with the C-terminal region of the HOPS subunit Vps39 via its RUN domain. Further biochemical and structural studies will be needed to understand the basis of this interaction.
Members of the Atg8 family undergo conjugation to phosphatidylethanolamine and this lipidation modification anchors these ubiquitin-like proteins to the inner and outer membranes of autophagosomes.Citation76 Recent imaging studies in mammalian cells revealed that the Atg8 conjugation system, more specifically the ATG3 E2-like enzyme, is important for the degradation of the autophagosome inner membrane after fusion with lysosomes.Citation77 Apart from binding and cooperating with tethering factors, Atg8 family members may participate in other aspects of autophagosome-lysosome fusion. In vitro synthetic liposomes-based studies showed that yeast Atg8 and mammalian LC3 and GABARAP proteins are capable of mediating membrane tethering and hemifusion.Citation78–83 More recently, it was demonstrated that mammalian LC3B and GABARAP show distinct membrane curvature-dependent tethering activities with LC3B more efficeHowever, it remains to be confirmed that the intrinsic membrane modulation properties allow Atg8 proteins to directly mediate autophagosome-lysoosme/vacuole fusion.
RAB and RAB regulators
The highly conserved Rab family of proteins are the master regulators of intracellular transport in eukaryotic cells. These small GTPases function as molecular switches by cycling between an inactive, cytoplasmic GDP-bound state and an active, membrane-bound GTP-bound state.Citation84 Active Rabs localize to unique membrane surfaces and control transport by recruiting specialized effectors to their sites of action.Citation85 Within the Rab family, Rab7 and its homologue yeast Ypt7 are required for autophagosome-lysosome/late endosome fusion.Citation86 Early X-ray crystallographic analysis of yeast Ypt7 and subsequent structural studies of Rab7 in complex with different effectors showed that Rab7/Ypt7 adopts a similar fold as other Rabs/Ypt proteins but with slight conformational differences in the Switch I and II regions, the N-terminal and C-terminal regions, as well as several loops that define binding specificity to effectors.Citation87,Citation88 Like all Rabs, the spatialtemporal activation and thereby membrane localization of Rab7/Ypt7 is tightly controlled by guanine nucleotide exchange factors (GEFs), a sequence divergent family of proteins and protein complexes that facilitate GDP to GTP exchange of their cognate Rabs.Citation90,Citation89 The bona fide Rab7/Ypt7 GEF is the evolutionarily conserved heterodimeric Mon1-Ccz1, a member of the Tri Longin Domain (TLD) Rab GEF family. This complex is a critical component of the Rab cascade that controls controlling endosome maturation. Mon1-Ccz1 is recruited by the early endosome-localized Rab5 and PI(3)P and this subsequently displaces Rabex5 (Rab5 GEF) from endosomal membrane to stop positive feedback loop of Rab5 activation.Citation91 Mon1-Ccz1 then recruits and activates Rab7 to drive late endosome maturation and fusion events.
In autophagy, Mon1-Ccz1 is recruited to autophagosomes in a Rab5-independent fashion likely through interaction between the conserved LIR motif at the C-terminus Ccz1 C-terminus and Atg8 family of proteins that are anchored to the outer membrane of autophagosomes.Citation92 Recent structural studies have generated insights into how Mon1-Ccz1 activates Rab7/Ypt7 and potentially gets recruited to membrane surfaces. The crystal structure of the catalytic core composed of LD1 (Longin domain 1) of Mon1 and the LD1 of Ccz1 in complex with Ypt7 revealed how the switch regions of Ypt7 are remodelled by Mon1-Ccz1 to open up the nucleotide-binding pocket to promote displacement of bound nucleotideCitation93 (). Regions outside of the core are believed to mediate membrane recruitment of Mon1-Ccz1. The recently reported cryo-EM structure of the full fungal Mon1-Ccz1complex from Chaetomium thermophilum showed that both subunits contain 3 Longin domains (LS) arranged in a triangular fashion, with LD1 and LD3 from the two subunits projected towards one another to mediate.Citation94 This structural model also showed that the additional LDs did not affect the configuration of the LD1-LD1 catalytic core. Analysis of the electrostatic potential of the surface of the complex led to the identification of a basic patch located in LD2 and LD3 of Mon1 that may serve as the PIP-containing membrane binding region of Mon1-Ccz1.
Figure 3. Rab7 regulator and its activator. Rab7 plays a key role in mediating late endosomal trafficking and autophagy and has been shown to localize to both the lysosome and the autophagosome to recruit different effectors. Rab7 is activated by a GEF which promotes displacement of GDP and loading of GTP. The GEF for Rab7 is the heterodimeric Mon1-Ccz1 complex. The crystal structure of the catalytic core Mon1-Ccz1 in complex with Rab7-orthologue Ypt7 from Thermochaetoides thermophila revealed how this GEF to promote nucleotide exchange (Mon1 in green, Ccz1 in gold, Ypt7 in blue). Mon1-Ccz1 in higher eukaryotes contain an additional component called RMC1 or Bulli. The cryo-EM structure of Drosophila melanogaster Mon1-Ccz1-Rmc1 complex revealed the third component adopts a leg-like architecture and binds to a region opposite to the Rab7 substrate binding site.
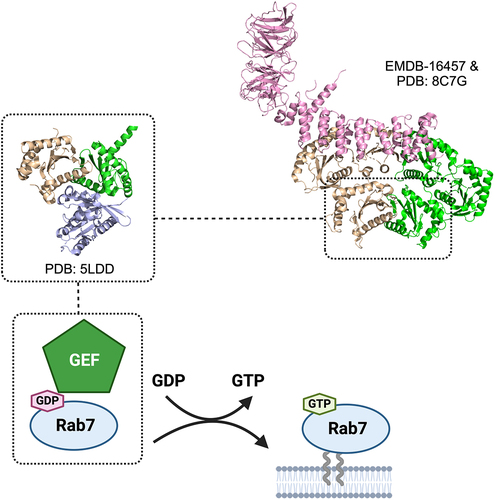
Interestingly, Mon1-Ccz1 in higher eukaryotes contain an additional non-TLD subunit called RMC1 or Bulli. The high-resolution structure of the Drosophila melanogaster Mon1-Ccz1-Rmc1 complex was reported by two separate research groups very recentlyCitation95,Citation96 (). Rmc1 was found to adopt a leg-like architecture composed of an N-terminal beta-propeller foot domain resembling the WD40 domain of yeast Atg18 but lacks the lipid binding motif and a C-terminal alpha-solenoid shin domain. Although Rmc1 binds to the opposite surface of the Rab7 substrate binding site and does not alter the structural configuration of catalytic core, the LD2 and LD2 domains of Mon1 and Ccz1 were observed to undergo significant conformational changes to accommodate the binding of the third subunit.
Summary
The identification of key components of the fusion machinery has generated a framework for dissecting the molecular mechanism of the autophagosome-lysosome/vacuole fusion process critical to the specific and efficient execution of autophagy. While many of the players involved in the autophagy fusion process are similar to (eg. Rabs, SNAREs) or even sometimes shared with (eg. HOPS) other intracellular transport pathways, unique factors have been uncovered that may function to provide an additional level of regulation of this process (eg. EPG5). Classical X-ray crystallographic-based studies have produced high-resolution snapshots of the human STX17-SNAP29-VAMP8 trans-SNARE complex central to autophagosome-lysosome fusion as well as a presumed intermediate containing fungal Vps33, the SM component of HOPS, in complex with two vacuolar SNAREs, and the fungal Mon1-Ccz1 catalytic core in complex with Ypt7. Further studies are still required to delineate the structural dynamics of these fusion components. The advent of single-particle cryo-EM and machine learning-based structural modeling have greatly facilitated structural investigations of all types of proteins and protein complexes. The recently reported molecular structure of yeast HOPS is a testament to the state of these cutting-edge technologies and illustrates how high-quality structural information can be obtained from a previously intractable dynamic assembly. We anticipate integrated structural approaches combining conventional X-ray crystallography, cryo-EM, and AlphaFold2 will produce exciting data to further our knowledge of the molecular structures and dynamics of other components of the autophagosome-lysosome/vacuole machinery. Future studies will also focus on delineating if, when, and how the different components are recruited to the fusion site and how they work collaboratively with one another to ensure the specificity and efficiency of fusion.
Acknowledgments
J.D. was supported by the NIH (R35GM128837). C.K.Y. was supported by a project grant (PJT-168907) from CIHR. Q. Z. was supported by grants from NSFC (91754205 and M-1040).
Additional information
Funding
References
- Zhao YG, Codogno P, Zhang H. Machinery, regulation and pathophysiological implications of autophagosome maturation. Nat Rev Mol Cell Biol. 2021;22:733–20.
- Zhao YG, Zhang H. Autophagosome maturation: an epic journey from the ER to lysosomes. J Cell Biol. 2019;218:757–770.
- Corona AK, Jackson WT. Finding the middle ground for autophagic fusion requirements. Trends Cell Biol. 2018;28:869–881.
- Lőrincz P, Juhász G. Autophagosome-lysosome fusion. J Mol Biol. 2020;432:2462–2482.
- Nakamura S, Yoshimori T. New insights into autophagosome-lysosome fusion. J Cell Sci. 2017;130:1209–1216.
- Tian X, Teng J, Chen J. New insights regarding SNARE proteins in autophagosome-lysosome fusion. Autophagy. 2021;17:2680–2688.
- Südhof TC, Rothman JE. Membrane fusion: grappling with SNARE and SM proteins. Science. 2009;323:474–477.
- Söllner T, Whiteheart SW, Brunner M, et al. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362:318–324.
- Fasshauer D, Sutton RB, Brunger AT, et al. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc Natl Acad Sci U S A. 1998;95:15781–15786.
- Lamb CA, Yoshimori T, Tooze SA. The autophagosome: origins unknown, biogenesis complex. Nat Rev Mol Cell Biol. 2013;14:759–774.
- Kloepper TH, Kienle CN, Fasshauer D. An elaborate classification of SNARE proteins sheds light on the conservation of the eukaryotic endomembrane system. Mol Biol Cell. 2007;18:3463–3471.
- Sutton RB, Fasshauer D, Jahn R, et al. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395:347–353.
- Tian Z, Gong J, Crowe M, et al. Biochemical studies of membrane fusion at the single-particle level. Prog Lipid Res. 2019;73:92–100.
- Brunger AT, Cipriano DJ, Diao J. Towards reconstitution of membrane fusion mediated by SNAREs and other synaptic proteins. Crit Rev Biochem Mol Biol. 2015;50:231–241.
- Jahn R, Scheller RH. SNAREs–engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–643.
- Furuta N, Fujita N, Noda T, et al. Combinational soluble N-ethylmaleimide-sensitive factor attachment protein receptor proteins VAMP8 and Vti1b mediate fusion of antimicrobial and canonical autophagosomes with lysosomes. Mol Biol Cell. 2010;21:1001–1010.
- Fader CM, Sánchez DG, Mestre MB, et al. TI-VAMP/VAMP7 and VAMP3/cellubrevin: two v-SNARE proteins involved in specific steps of the autophagy/multivesicular body pathways. Biochim Biophys Acta. 2009;1793:1901–1916.
- Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151:1256–1269.
- Takáts S, Nagy P, Varga Á, et al. Autophagosomal Syntaxin17-dependent lysosomal degradation maintains neuronal function in Drosophila. J Cell Biol. 2013;201(4):531–539.
- Diao J, Liu R, Rong Y, et al. ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature. 2015;520:563–566.
- Bu B, Tian Z, Li D, et al. Double-transmembrane domain of SNAREs decelerates the fusion by increasing the protein-lipid mismatch. J Mol Biol. 2023;435:168089.
- Matsui T, Jiang P, Nakano S, et al. Autophagosomal YKT6 is required for fusion with lysosomes independently of syntaxin 17. J Cell Biol. 2018;217:2633–2645.
- Bas L, Papinski D, Licheva M, et al. Reconstitution reveals Ykt6 as the autophagosomal SNARE in autophagosome-vacuole fusion. J Cell Biol. 2018;217:3656–3669.
- Takáts S, Glatz G, Szenci G, et al. Non-canonical role of the SNARE protein Ykt6 in autophagosome-lysosome fusion. PLoS Genet. 2018;14:e1007359.
- Malmersjö S, Di Palma S, Diao J, et al. Phosphorylation of residues inside the SNARE complex suppresses secretory vesicle fusion. EMBO J. 2016;35:1810–1821.
- Wang L, Diao J. VAMP8 phosphorylation regulates lysosome dynamics during autophagy. Autophagy Rep. 2022;1:79–82.
- Guo B, Liang Q, Li L, et al. O-GlcNAc-modification of SNAP-29 regulates autophagosome maturation. Nat Cell Biol. 2014;16:1215–1226.
- Wang P, Lazarus BD, Forsythe ME, et al. O-GlcNAc cycling mutants modulate proteotoxicity in Caenorhabditis elegans models of human neurodegenerative diseases. Proc Natl Acad Sci U S A. 2012;109:17669–17674.
- Pellegrini FR, De Martino S, Fianco G, et al. Blockage of autophagosome-lysosome fusion through SNAP29 O-GlcNAcylation promotes apoptosis via ROS production. Autophagy. 2023;19:2078–2093.
- Chen Q, Hao M, Wang L, et al. Prefused lysosomes cluster on autophagosomes regulated by VAMP8. Cell Death Dis. 2021;12:939.
- Huang H, Ouyang Q, Zhu M, et al. mTOR-mediated phosphorylation of VAMP8 and SCFD1 regulates autophagosome maturation. Nat Commun. 2021;12:6622.
- Shen Q, Shi Y, Liu J, et al. Acetylation of STX17 (syntaxin 17) controls autophagosome maturation. Autophagy. 2021;17:1157–1169.
- Kumar S, Gu Y, Abudu YP, et al. Phosphorylation of syntaxin 17 by TBK1 controls autophagy initiation. Dev Cell. 2019;49:130–144.e6.
- Gao J, Kurre R, Rose J, et al. Function of the SNARE Ykt6 on autophagosomes requires the Dsl1 complex and the Atg1 kinase complex. EMBO Rep. 2020;21:e50733.
- Barz S, Kriegenburg F, Henning A, et al. Atg1 kinase regulates autophagosome-vacuole fusion by controlling SNARE bundling. EMBO Rep. 2020;21:e51869.
- Sánchez-Martín P, Kriegenburg F, Alves L, et al. ULK1-mediated phosphorylation regulates the conserved role of YKT6 in autophagy. J Cell Sci. 2023;136:jcs260546.
- Herrmann JM, Spang A. Intracellular parcel service: current issues in intracellular membrane trafficking. Methods Mol Biol. 2015;1270:1–12.
- Yu I-M, Hughson FM. Tethering factors as organizers of intracellular vesicular traffic. Annu Rev Cell Dev Biol. 2010;26:137–156.
- Stanton AE, Hughson FM. The machinery of vesicle fusion. Curr Opin Cell Biol. 2023;83:102191.
- Nickerson DP, Brett CL, Merz AJ. Vps-C complexes: gatekeepers of endolysosomal traffic. Curr Opin Cell Biol. 2009;21:543–551.
- Takáts S, Pircs K, Nagy P, et al. Interaction of the HOPS complex with Syntaxin 17 mediates autophagosome clearance in Drosophila. Mol Biol Cell. 2014;25:1338–1354.
- Jiang P, Nishimura T, Sakamaki Y, et al. The HOPS complex mediates autophagosome-lysosome fusion through interaction with syntaxin 17. Mol Biol Cell. 2014;25:1327–1337.
- Solinger JA, Spang A. Tethering complexes in the endocytic pathway: CORVET and HOPS. FEBS J. 2013;280:2743–2757.
- Orr A, Song H, Rusin SF, et al. HOPS catalyzes the interdependent assembly of each vacuolar SNARE into a SNARE complex. Mol Biol Cell. 2017;28:975–983.
- McEwan DG, Popovic D, Gubas A, et al. PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol Cell. 2015;57:39–54.
- Stroupe C, Collins KM, Fratti RA, et al. Purification of active HOPS complex reveals its affinities for phosphoinositides and the SNARE Vam7p. EMBO J. 2006;25:1579–1589.
- Jia R, Guardia CM, Pu J, et al. BORC coordinates encounter and fusion of lysosomes with autophagosomes. Autophagy. 2017;13:1648–1663.
- Schleinitz A, Pöttgen L-A, Keren-Kaplan T, et al. Consecutive functions of small GTPases guide HOPS-mediated tethering of late endosomes and lysosomes. Cell Rep. 2023;42:111969.
- Baker RW, Jeffrey PD, Hughson FM. Crystal structures of the Sec1/Munc18 (SM) protein Vps33, alone and bound to the homotypic fusion and vacuolar protein sorting (HOPS) subunit Vps16*. PLoS One. 2013;8:e67409.
- Graham SC, Wartosch L, Gray SR, et al. Structural basis of Vps33A recruitment to the human HOPS complex by Vps16. Proc Natl Acad Sci U S A. 2013;110:13345–13350.
- Baker RW, Jeffrey PD, Zick M, et al. A direct role for the Sec1/Munc18-family protein Vps33 as a template for SNARE assembly. Science. 2015;349:1111–1114.
- Behrmann H, Lürick A, Kuhlee A, et al. Structural identification of the Vps18 β-propeller reveals a critical role in the HOPS complex stability and function. J Biol Chem. 2014;289:33503–33512.
- Bröcker C, Kuhlee A, Gatsogiannis C, et al. Molecular architecture of the multisubunit homotypic fusion and vacuole protein sorting (HOPS) tethering complex. Proc Natl Acad Sci U S A. 2012;109:1991–1996.
- Chou H-T, Dukovski D, Chambers MG, et al. CATCHR, HOPS and CORVET tethering complexes share a similar architecture. Nat Struct Mol Biol. 2016;23:761–763.
- Shvarev D, Schoppe J, König C, et al. Structure of the HOPS tethering complex, a lysosomal membrane fusion machinery. Elife. 2022;11:e80901.
- Zhang S, Tong M, Zheng D, et al. C9orf72-catalyzed GTP loading of Rab39A enables HOPS-mediated membrane tethering and fusion in mammalian autophagy. Nat Commun. 2023;14:6360.
- Tian Y, Li Z, Hu W, et al. C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell. 2010;141:1042–1055.
- Wang Z, Miao G, Xue X, et al. The Vici syndrome protein EPG5 is a Rab7 effector that determines the fusion specificity of autophagosomes with late endosomes/lysosomes. Mol Cell. 2016;63:781–795.
- Cullup T, Kho AL, Dionisi-Vici C, et al. Recessive mutations in EPG5 cause Vici syndrome, a multisystem disorder with defective autophagy. Nat Genet. 2013;45:83–87.
- Nam S-E, Cheung YW, Nguyen TN, et al. Insights on autophagosome-lysosome tethering from structural and biochemical characterization of human autophagy factor EPG5. Commun Biol. 2021;4:291.
- Fan W, Nassiri A, Zhong Q. Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L). Proc Natl Acad Sci U S A. 2011;108:7769–7774.
- Matsunaga K, Saitoh T, Tabata K, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–396.
- Matsunaga K, Morita E, Saitoh T, et al. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J Cell Biol. 2010;190:511–521.
- Itakura E, Kishi C, Inoue K, et al. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372.
- Zhong Y, Wang QJ, Li X, et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–476.
- Sun Q, Fan W, Zhong Q. Regulation of Beclin 1 in autophagy. Autophagy. 2009;5:713–716.
- Hamasaki M, Furuta N, Matsuda A, et al. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013;495:389–393.
- Chen D, Fan W, Lu Y, et al. A mammalian autophagosome maturation mechanism mediated by TECPR1 and the Atg12-Atg5 conjugate. Mol Cell. 2012;45:629–641.
- Kim JH, Hong SB, Lee JK, et al. Insights into autophagosome maturation revealed by the structures of ATG5 with its interacting partners. Autophagy. 2015;11:75–87.
- Florey O. TECPR1 helps bridge the CASM during lysosome damage. EMBO J. 2023;42:e115210.
- Corkery DP, Castro-Gonzalez S, Knyazeva A, et al. An ATG12-ATG5-TECPR1 E3-like complex regulates unconventional LC3 lipidation at damaged lysosomes. EMBO Rep. 2023;24:e56841.
- Boyle KB, Ellison CJ, Elliott PR, et al. TECPR1 conjugates LC3 to damaged endomembranes upon detection of sphingomyelin exposure. EMBO J. 2023;42:e113012.
- Kaur N, de la Ballina LR, Haukaas HS, et al. TECPR1 is activated by damage-induced sphingomyelin exposure to mediate noncanonical autophagy. EMBO J. 2023;42:e113105.
- Ebner P, Poetsch I, Deszcz L, et al. The IAP family member BRUCE regulates autophagosome-lysosome fusion. Nat Commun. 2018;9:599.
- Marwaha R, Arya SB, Jagga D, et al. The Rab7 effector PLEKHM1 binds Arl8b to promote cargo traffic to lysosomes. J Cell Biol. 2017;216:1051–1070.
- Shpilka T, Weidberg H, Pietrokovski S, et al. Atg8: an autophagy-related ubiquitin-like protein family. Genome Biol. 2011;12:226.
- Tsuboyama K, Koyama-Honda I, Sakamaki Y, et al. The ATG conjugation systems are important for degradation of the inner autophagosomal membrane. Science. 2016;354:1036–1041.
- Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–178.
- Weidberg H, Shpilka T, Shvets E, et al. LC3 and GATE-16 N termini mediate membrane fusion processes required for autophagosome biogenesis. Dev Cell. 2011;20:444–454.
- Nair U, Jotwani A, Geng J, et al. SNARE proteins are required for macroautophagy. Cell. 2011;146:290–302.
- Yang A, Li Y, Pantoom S, et al. Semisynthetic lipidated LC3 protein mediates membrane fusion. Chembiochem. 2013;14:1296–1300.
- Wu F, Watanabe Y, Guo XY, et al. Structural basis of the differential function of the two C. elegans Atg8 homologs, LGG-1 and LGG-2, in autophagy. Mol Cell. 2015;60:914–929.
- Landajuela A, Hervás J, Antón Z, et al. Lipid geometry and bilayer curvature modulate LC3/GABARAP-mediated model autophagosomal elongation. Biophys J. 2016;110:411–422.
- Hutagalung AH, Novick PJ. Role of Rab GTPases in membrane traffic and cell physiology. Physiol Rev. 2011;91:119–149.
- Goody RS, Müller MP, Wu Y-W. Mechanisms of action of Rab proteins, key regulators of intracellular vesicular transport. Biol Chem. 2017;398:565–575.
- Kuchitsu Y, Fukuda M. Revisiting Rab7 functions in mammalian autophagy: rab7 knockout studies. Cells. 2018;7:215.
- Rak A, Pylypenko O, Niculae A, et al. Structure of the Rab7:REP-1 complex: insights into the mechanism of Rab prenylation and choroideremia disease. Cell. 2004;117:749–760.
- Wu M, Wang T, Loh E, et al. Structural basis for recruitment of RILP by small GTPase Rab7. EMBO J. 2005;24:1491–1501.
- Blümer J, Rey J, Dehmelt L, et al. RabGEFs are a major determinant for specific Rab membrane targeting. J Cell Biol. 2013;200:287–300.
- Müller MP, Goody RS. Molecular control of Rab activity by GEFs, GAPs and GDI. Small GTPases. 2018;9:5–21.
- Borchers A-C, Langemeyer L, Ungermann C. Who’s in control? Principles of Rab GTPase activation in endolysosomal membrane trafficking and beyond. J Cell Biol. 2021;220:e202105120.
- Gao J, Langemeyer L, Kümmel D, et al. Molecular mechanism to target the endosomal Mon1-Ccz1 GEF complex to the pre-autophagosomal structure. Elife. 2018;7:e31145.
- Kiontke S, Langemeyer L, Kuhlee A, et al. Architecture and mechanism of the late endosomal Rab7-like Ypt7 guanine nucleotide exchange factor complex Mon1-Ccz1. Nat Commun. 2017;8:14034.
- Klink BU, Herrmann E, Antoni C, et al. Structure of the Mon1-Ccz1 complex reveals molecular basis of membrane binding for Rab7 activation. Proc Natl Acad Sci U S A. 2022;119:e2121494119.
- Herrmann E, Schäfer J-H, Wilmes S, et al. Structure of the metazoan Rab7 GEF complex Mon1-Ccz1-Bulli. Proc Natl Acad Sci U S A. 2023;120:e2301908120.
- Yong X, Jia G, Liu Z, et al. Cryo-EM structure of the Mon1-Ccz1-RMC1 complex reveals molecular basis of metazoan RAB7A activation. Proc Natl Acad Sci U S A. 2023;120:e2301725120.

