Figures & data
Figure 1. Selective oxomer formation between FIH and IκBβ. (A) Immunoblot analysis of the oxomer formation between ectopically expressed FIH-V5 and V5-IκBβ in HEK293 cells, using FIH-OTUB1 oxomer formation via ectopic expression of FIH-V5 and FLAG-OTUB1 as control. (B) Immunoblot analysis of FIH-IκBβ oxomer formation in HepG2, Hep3B and MCF7 cells with ectopic FIH-V5 and IκBβ-FLAG expression. (C) Immunoprecipitation of endogenous IκBβ from HEK293 cells. A signal was detected at a molecular weight consistent with a FIH-IκBβ oxomer (highlighted by *). This complex was absent in the presence of the hydroxylase inhibitor DMOG (1 mM for 24 h) and in FIH knockout (KO) cells. (D) Immunoblot analysis of oxomer formation between FIH-V5 and the Indicated members of the IκB protein family following transient transfection of HEK293 cells with corresponding vectors for ectopic expression. Predicted molecular weights of putative FIH oxomers (without post-translational protein modifications): FIH-IκBα, 76 kDa; FIH-IκBβ, 78 kDa; FIH-Bcl-3, 88 kDa; FIH-IκBδ, 90 kDa; FIH-IκBε, 78 kDa; FIH-p105, 145 kDa; FIH-p100, 137 kDa. Data are representative of three (A, B, D) and two (C) independent experiments. F, FIH-V5; I, V5-IκBβ; F-O, FIH-OTUB1 oxomer; F-I, FIH-IκBβ oxomer; interm., intermediate; exp, exposure; DMOG, dimethyloxalylglycine; ns, non-specific.
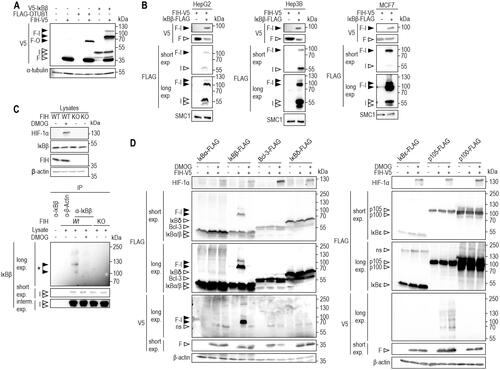
Figure 2. FIH-IκBβ oxomer formation depends on FIH enzymatic activity and is highly hypoxia-sensitive. (A) FIH-IκBβ oxomer formation is prevented by hypoxia (0.2% O2), DMOG (1 mM) and DFX (0.1 mM), but not by the PHD-selective FG-4592 (0.1 mM) or the FIH-selective DM-NOFD (1 mM). Treatments were performed for 16 h in HEK293 cells. (B, C) Genetically inactivated FIH (H199A or L340R loss of function mutations) prevented FIH-IκBβ oxomer formation. (D) Hypoxia sensitivity of HIF-1α stabilization and FIH-IκBβ oxomer formation after 24 h at the indicated oxygen levels. (E) Quantification of the results shown in (D) and calculation of the corresponding oxygen sensitivities. Data are shown as mean ± SEM. F, FIH-V5; I, IκBβ-FLAG; Nx, normoxia; Hx, hypoxia (0.2% O2); WT, wild-type. Data are representative of three (A-C) and four (D, E) independent experiments.
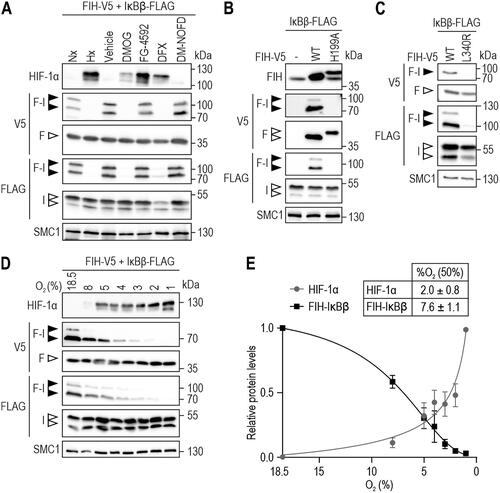
Figure 3. FIH-IκBβ oxomer stability. (A) Immunoblot analysis of the FIH-IκBβ oxomer upon treatment of cell lysates with the indicated reducing agents followed by incubation for 5 min at 95 °C. (B) FIH-IκBβ oxomer formation in HEK293 cells following the indicated transfection and treatment with the proteasome inhibitor MG132 (10 μM). (C) HEK293 cells were transfected with vectors encoding FIH and IκBβ for 22 h to form the FIH-IκBβ oxomer. Subsequently, oxomer stability was analyzed in hypoxia (0.2% O2), inhibiting FIH and thus preventing any additional oxomer formation. (D) Quantification of the results shown in (C), presented as mean ± SEM. F, FIH-V5; I, IκBβ-FLAG; F-I, FIH-IκBβ oxomer; DTT, dithiothreitol; interm., intermediate; exp., exposure. *P < 0.05 by Student’s t test. Data are representative of three independent experiments.
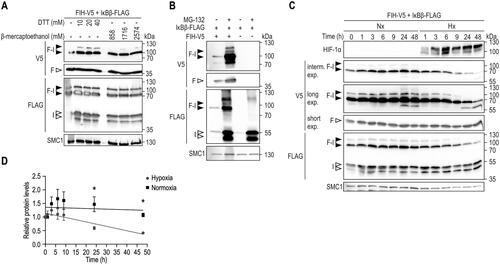
Figure 4. Analysis of potential IκBβ amino acid residues as substrates for FIH-IκBβ oxomer formation. (A) Overlay of protein structures of IκBα (shown in cyan, identifier AF-P25963-F1) and IκBβ (shown in green, identifier AF-Q15653-F1) predicted by AlphaFold Protein Structure Database. Asparagine residues of IκBα previously demonstrated to be hydroxylated by FIHCitation17 are indicated in dark blue, all asparagine residues of IκBβ are highlighted in red. An aspartic acid residue that had previously been found to be oxidized is highlighted in magenta.Citation12 (B) Separate point mutations were introduced in IκBβ-FLAG, changing each asparagine residue and the highlighted aspartic acid residue to alanine. Oxomer formation was assessed in HEK293 cells by immunoblotting. (C) Analysis of oxomer formation between FIH and truncated IκBβ (ARD 1–3, 1–166 amino acids; ARD 4–6, 167–356 amino acids of full length IκBβ). (D) Analysis of oxomer formation between FIH and the indicated C-terminally truncated IκBβ fragments. (E) Analysis of oxomer formation between FIH and IκBβ containing the indicated amino acid deletions. Amino acids identified to be involved in oxomer formation (122–126) are highlighted in red. F, FIH-V5; I, IκBβ-FLAG; WT, wild-type; ARD, ankyrin repeat domain; *FIH-IκBβ oxomer with truncated IκBβ forms; trunc., truncated; exp., exposure. Predicted molecular weights of proteins without tags (kDa): ARD 1–3: 21, ARD 4–6: 25; predicted molecular weights of oxomers without tags (kDa): FIH-IκBβ ARD 1–3: 61, FIH-IκBβ ARD 4–6: 65. Data are representative of three (B), one (C and E) and two (D) independent experiments.
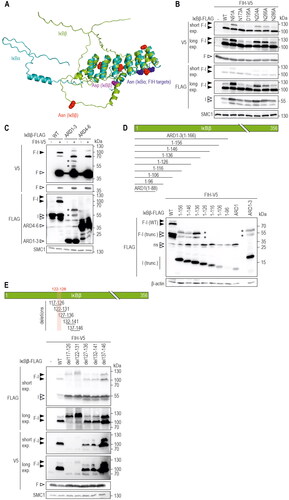
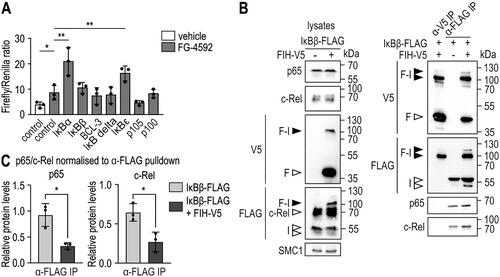
Figure 5. FIH-IκBβ oxomer formation prevents IκBβ from binding NF-κB subunits. (A) HEK293 cells were transiently cotransfected with a HIF-driven luciferase plasmid (pH3SVL) together with a constitutive Renilla luciferase plasmid (pRL-SV40) as well as plasmids expressing the indicated IκB protein family member 24 h prior to treatment. FG-4592 treatment was applied for 24 h in a concentration of 0.1 mM. Efficient transfection and FG-4592 treatment were determined by immunoblotting (Figure S5). Significance was assessed by two-way ANOVA with Tukey’s correction; *P < 0.05, **P < 0.01. (B) Immunoprecipitation (IP) analysis of the interaction of IκBβ and the FIH-IκBβ oxomer with the NF-κB subunits c-Rel and p65. (C) Quantification of the results shown in B. The p65 and c-Rel levels were normalized to the levels of total bait/FLAG pulldown (IκBβ or IκBβ + oxomer), respectively. Statistical analysis was performed by Student’s t test. *P < 0.05; F, FIH-V5; I, IκBβ-FLAG. Data are shown as mean ± SD. Data are representative of three independent experiments.
Supplemental Material
Download Zip (3.5 MB)Data availability statement
All underlying raw data have been made available on https://dataverse.harvard.edu/. The data has obtained the DOI 10.7910/DVN/XYXNPA and is accessible under https://dataverse.harvard.edu/privateurl.xhtml?token=29762520-1072-418d-873a-5a110caa04cf. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDECitation60 partner repository with the dataset identifier PXD047442.
