Figures & data
Figure 1. Artificial intelligence (AI) applications and tools in RNA-targeted small molecule drug discovery. Successful applications of AI tools in the various stages of drug discovery have been reported. CNN, convolutional neural network; DNN, deep neural network; GNN, graph neural network; Lasso, least absolute shrinkage and selection operator; LDA, linear discriminant analysis; LSTM, long short-term memory; MFFNN, multilayer feed-forward neural network; MLP, multilayer perceptron; MRF, Markov random fields; QSAR/QSPR, quantitative structure – activity/property relationship; SVM, support vector machine.
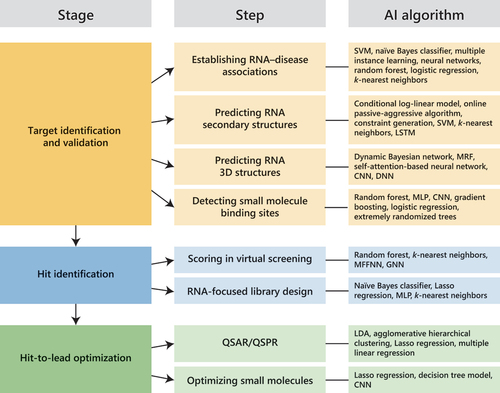
Table 1. Advantages and disadvantages of the ML and DL algorithms used in RNA-targeted small molecule drug discovery.
Table 2. Data repositories containing ncRNA – disease associations*.
Table 3. RNA secondary and tertiary structure prediction tools that employs AI methods*.
Figure 2. (a) Workflow for the DeepFoldRNA method [Citation48]. (b) Model of Saccharomyces cerevisiae P-site tRNA generated by DeepFoldRNA (green) superimposed with the native structure (blue, PDB ID: 6Q8Y). L-BFGS, limited-memory Broyden-Fletcher-Goldfarb-Shanno; RMSD, root mean square deviation.
![Figure 2. (a) Workflow for the DeepFoldRNA method [Citation48]. (b) Model of Saccharomyces cerevisiae P-site tRNA generated by DeepFoldRNA (green) superimposed with the native structure (blue, PDB ID: 6Q8Y). L-BFGS, limited-memory Broyden-Fletcher-Goldfarb-Shanno; RMSD, root mean square deviation.](/cms/asset/73b8fc00-86c3-429b-b158-fb013f8dc4d9/iedc_a_2313455_f0002_oc.jpg)
Figure 3. Chemical substructures enriched in selective binders in RNA-focused libraries. The Merck group adapted the (a) Automated ligand identification system as a method to screen small molecule libraries against RNAs [Citation87]. Using an ML approach and the results of their screening, they built an RNA-focused library and found (b) 12 substructures enriched in RNA binders. Likewise, the Schneekloth group built an RNA-focused library, ROBIN, from their previous (c) Small molecule microarray screening projects and identified (d) Ten substructures enriched in selective RNA binders [Citation88]. RP-HPLC, reversed-phase high-performance liquid chromatography.
![Figure 3. Chemical substructures enriched in selective binders in RNA-focused libraries. The Merck group adapted the (a) Automated ligand identification system as a method to screen small molecule libraries against RNAs [Citation87]. Using an ML approach and the results of their screening, they built an RNA-focused library and found (b) 12 substructures enriched in RNA binders. Likewise, the Schneekloth group built an RNA-focused library, ROBIN, from their previous (c) Small molecule microarray screening projects and identified (d) Ten substructures enriched in selective RNA binders [Citation88]. RP-HPLC, reversed-phase high-performance liquid chromatography.](/cms/asset/256e5079-279a-41ba-996b-ca34e50841bb/iedc_a_2313455_f0003_oc.jpg)
Figure 4. Application of AI in the optimization of small molecules that target (a) RNA hairpin 91 (red) in the ribosomal peptidyl transfer center of mycobacterium tuberculosis (PDB ID: 5O61). (b) Secondary structure of hairpin 91. (c) Chemical structures, ribosome activity, and inhibitory concentration (IC50) of the small molecules that were designed based on the insights obtained from the machine and deep learning approaches [Citation100].
![Figure 4. Application of AI in the optimization of small molecules that target (a) RNA hairpin 91 (red) in the ribosomal peptidyl transfer center of mycobacterium tuberculosis (PDB ID: 5O61). (b) Secondary structure of hairpin 91. (c) Chemical structures, ribosome activity, and inhibitory concentration (IC50) of the small molecules that were designed based on the insights obtained from the machine and deep learning approaches [Citation100].](/cms/asset/a45a3a48-7360-44e9-a66e-ab9d16e139e1/iedc_a_2313455_f0004_oc.jpg)
