Figures & data
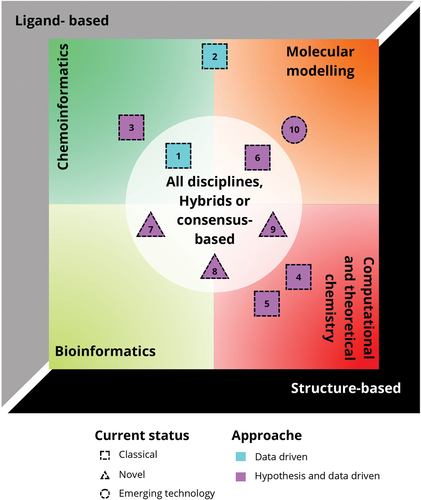
Figure 1. Timeline of automation of CADD. The X-axis represents the timeline of the most representative periods in CADD, and Y-axis illustrates the relative progress of automation in CADD. The continue line illustrates the events before 2024, while the dotted line illustrates the future events that CADD faces. Color coding is used to illustrate past (marine blue), emerging (purple), and future (cyan) events. Finally, the pill represents the current progress around automation in CADD.
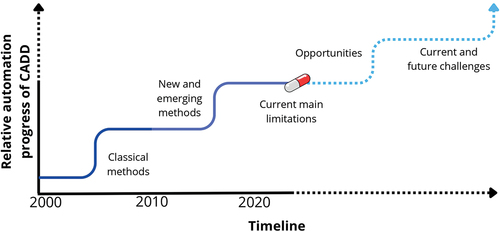
Figure 2. Different classifications of CADD and its role in modern DD. Classification method-based: Ligand-based, structure-based, and hybrids or consensus-based; classification theoretical discipline-based: chemoinformatics, molecular modeling, bioinformatics, or computational and theoretical chemistry; classification approach-based: data driven and hypothesis driven; Representative examples of methods used: (1) QSAR/QSPR modeling, (2) similarity searching, (3) chemical space analysis, (4) molecular docking, (5) molecular dynamics, (6) pharmacophore modeling, (7) machine or deep learning, (8) AI-QSAR, (9) generative or de novo design, and (10) quantum computing.