Figures & data
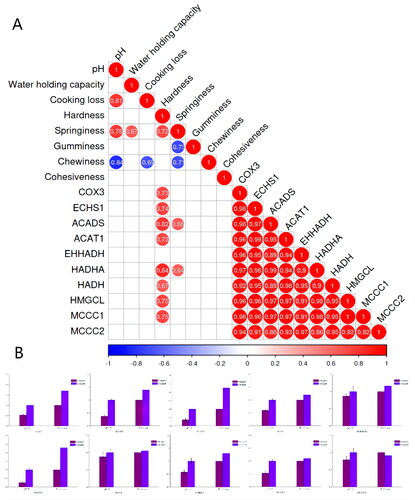
Table 1. Muscle physical and texture parameters of grass carp after 120-days feeding experiment.
Table 2. Summary statistics of transcriptome sequencing from muscle tissue between ordinary grass carp and the crisp carp group.
Figure 1. Volcano map and heat map of clustering of differentially expressed genes in Ctenopharyngodon idellus.
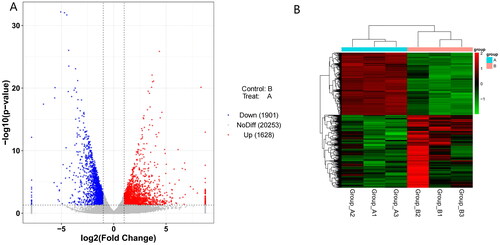
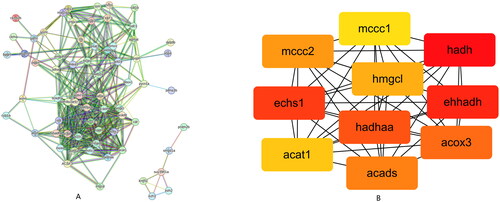
Figure 2. Classify and analysis according to gene function. (A) GO enrichment of top 20 up-regulated and down-regulated DEGs. (B) KEGG enrichment of top 20 up-regulated and down-regulated DEGs.
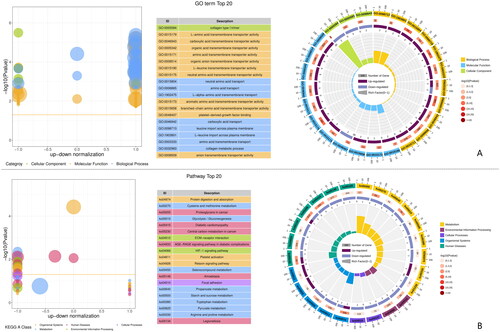
Table 3. Differentially expressed genes (DEGs) identified in the muscle tissue between ordinary grass carp and the crisp carp group (Top20).
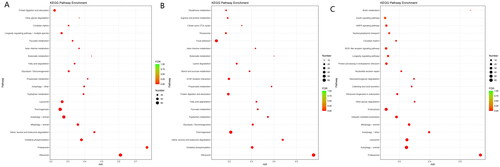
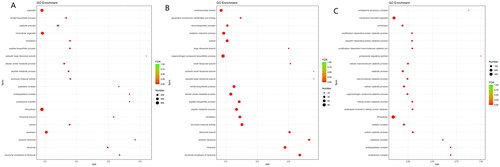
Figure 3. (A) GO enrichment of DEGs. (B) GO enrichment of up-regulated DEGs. (C) GO enrichment of down-regulated DEGs.
Supplemental Material
Download MS Excel (7.7 MB)Data availability statement
The data that support the fundings of this study are available from the corresponding author upon reasonable request.