Figures & data
Figure 1. Components of gut microbiota, their approximate size and abundance.
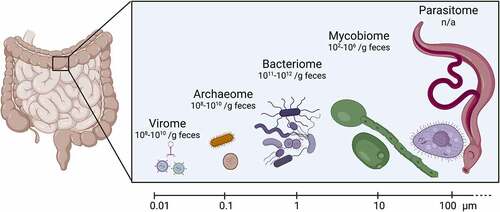
Figure 2. Effect of the gut virome on enteric bacterial infections.
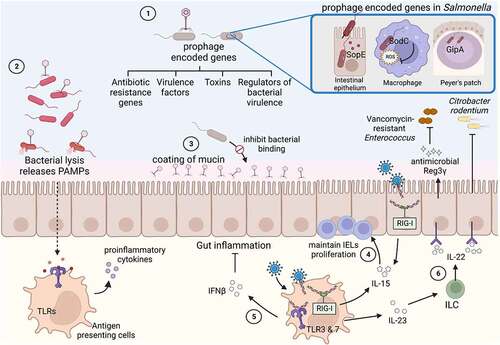
Figure 3. Effect of the gut archaeome on enteric bacterial infections.
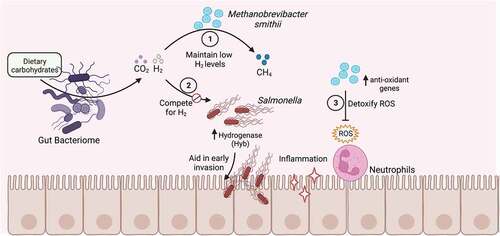
Figure 4. Direct interactions of gut fungi with enteric bacterial pathogens.
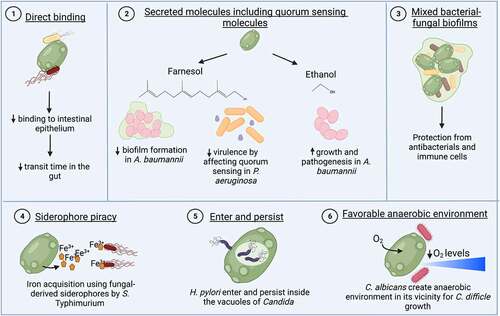
Figure 5. Indirect effects of the gut mycobiome on enteric bacterial infections.
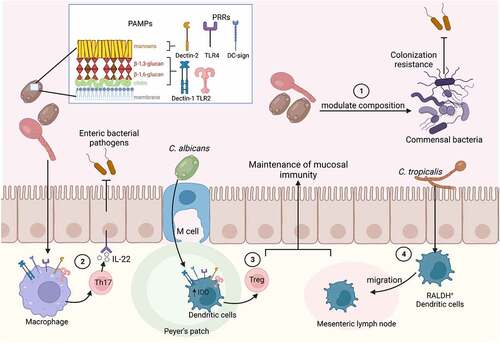
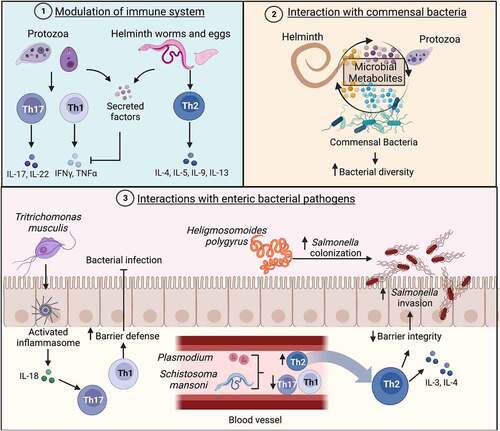
Figure 6. Effects of the gut parasitome on enteric bacterial infections.