Figures & data
Figure 1. Colibactin exacerbates DSS-induced colitis.
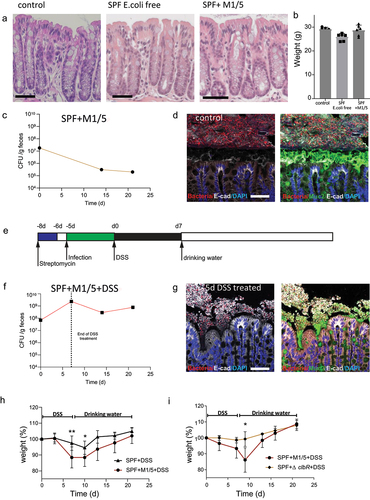
Figure 2. Colibactin increases the severity of tissue damage after DSS.
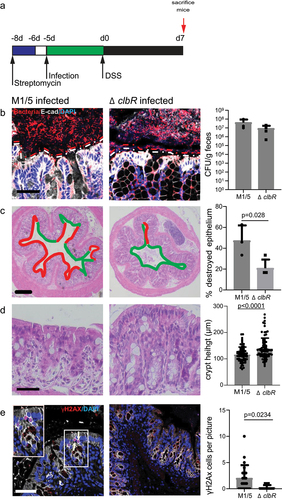
Figure 3. Colibactin delays tissue regeneration upon injury.
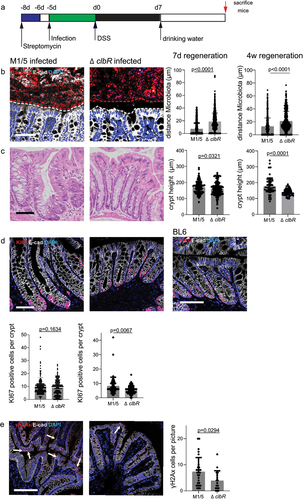
Figure 4. Infection with pks+ E. coli causes inflammation that resembles ulcerative colitis.
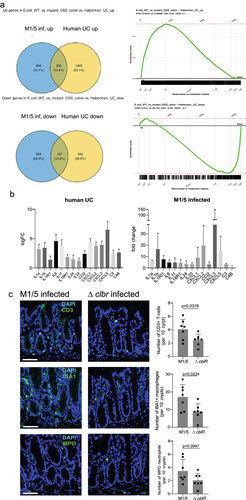
Figure 5. Epithelial changes seen in mice infected with WT M1/5 E. coli resemble those of human ulcerative colitis.
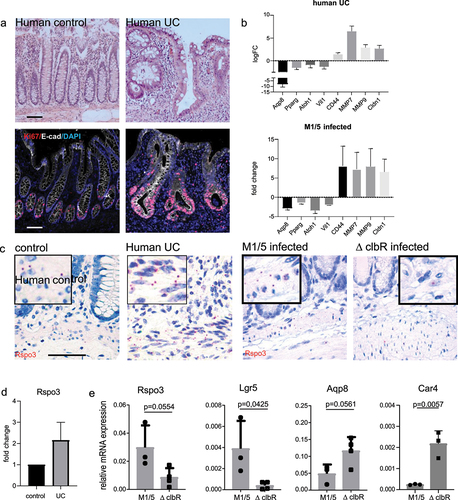
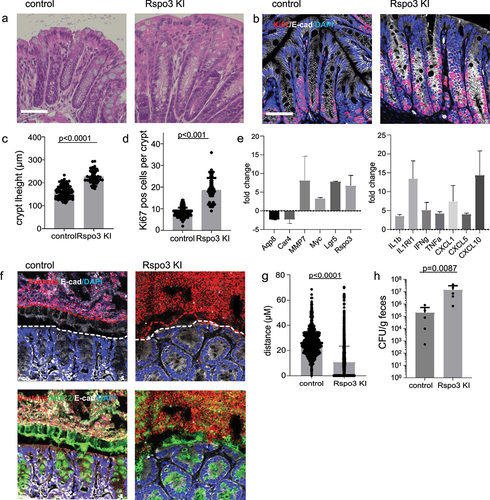
Figure 6. R-spondin 3 overexpression is sufficient to disrupt the mucosal barrier.
Data availability statement
Microarray data have been deposited in the gene expression omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) of the National Center for Biotechnology Information under accession number GSE205403, reviewer token: sxcrkgounnorlmd.
