Figures & data
Figure 1. Colonization Factors in gut commensal bacteria.
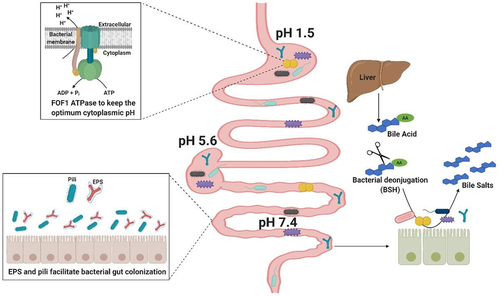
Table 1. Intracellular accumulation of glycogen reported in gut-related bacteria.
Figure 2. Glycogen metabolism pathways in gut commensal bacteria.
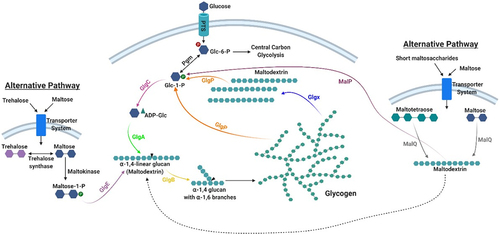
Table 2. Biosynthetic and degradative enzymes in the metabolism of glycogen.
Figure 3. Presence/Absence of predicted glycogen biosynthetic and degradative enzymes in gut bacteria.
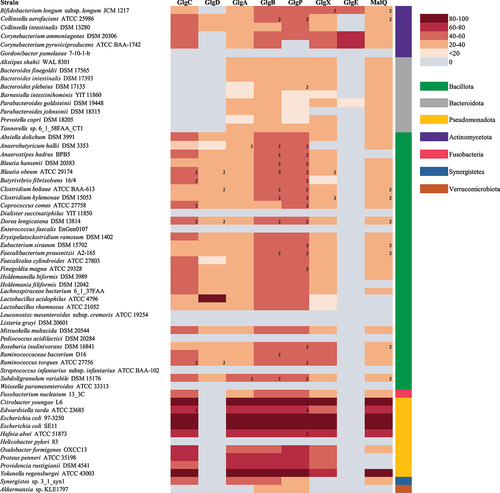
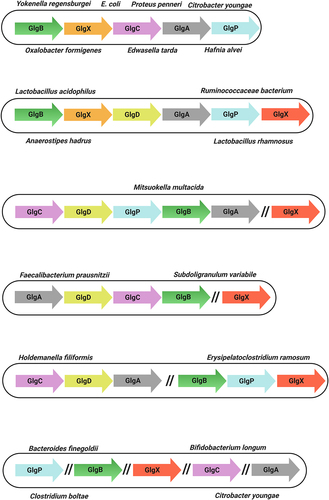
Figure 4. Genetic organization of glycogen genes in gut commensal bacteria.
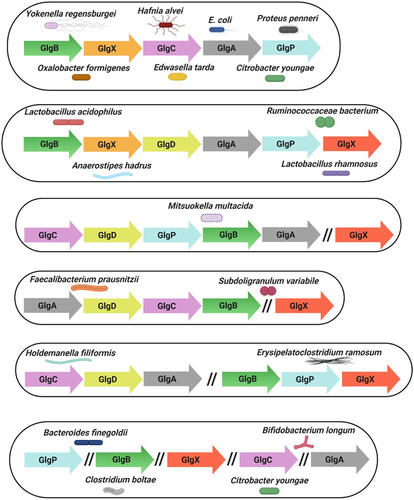
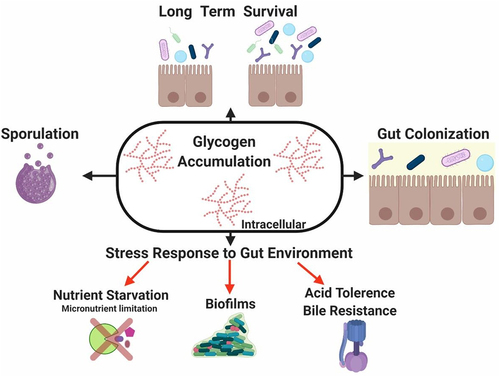
Figure 5. Role of glycogen metabolism in gut commensal bacteria.