Figures & data
Figure 1. Cellular and genomic changes during spermiogenesis. (a) Sperm undergo dramatic morphological differentiation from round haploid cells after meiosis to elongated nuclei with flagellar tails (top). Meanwhile, the genome first becomes haploid after meiosis, and then changes genomic architecture as histones are exchanged for protamines. Importantly, cells in meiosis I or II have homologous chromosomes or sister chromatids that provide 3 or 1 DNA templates, respectively, for potential DNA repair (bottom). Note that homologous recombination is not depicted in this cartoon. (b-d) Spermiogenesis contains 3 major challenges to genome integrity (see main text).
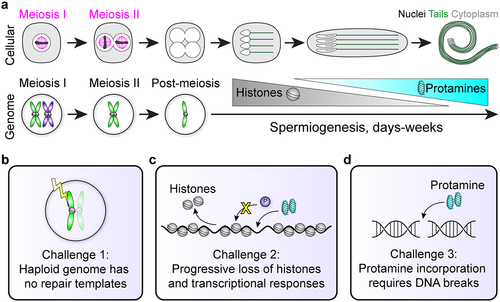
Figure 2. Potential solution to spermiogenesis challenges. (a) If developing sperm are damaged during spermiogenesis, how are they be detected and what is their fate? (b) We propose that a novel checkpoint could exist that does not rely on canonical transcription-based responses, nucleosomes and histone modifications, or a second DNA template.
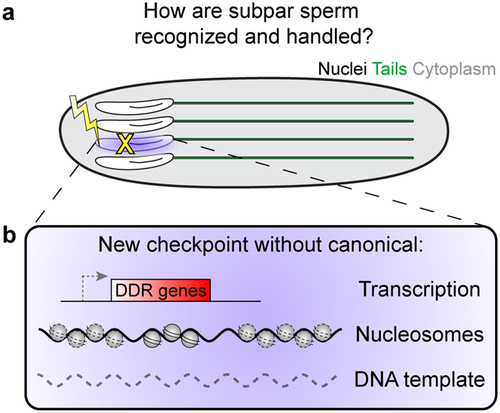
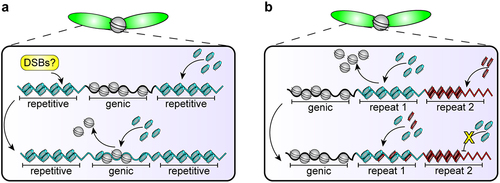
Figure 3. Hypothetical example of co-evolution between DNA sequences and protamines. (a) Perhaps protamines have preferential affinity for repetitive sequences, which could provide a signal to create DSBs safely at repetitive sequences so that genic sequences are not mutated but can incorporate protamines. (b) This preferential affinity could evolve to be sequence-specific such that the ‘wrong’ protamine-DNA pair cannot compact, causing interference with and ultimately failure of the histone-to-protamine transition.
Data availability statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
