Figures & data
Figure 1. Schematic illustration displaying the procedure of this study. (a) RNA-seq data were either downloaded from the GEO database or generated in-house. Samples without detailed experimental information or with no experimental replicates were excluded. (b) RNA-seq reads were mapped to the S. aureus pangenome reference and transcripts abundances were quantified using salmon software. (c) Expression matrix derived from the previous step was used as the input data for the ICA analysis pipeline. Resulting in M and a tables containing iModulon information. M table: Each column represents an independent component (IC), and each row contains the gene weights for each gene across each IC: Each column represents a sample, and each row contains the activity of each iModulon across all samples. (d) The M and a tables together with sample information were used as input for iModulon characterization using python package pymodulon. Components of iModulon containing both sRNA and genes were identified, and their activity under each experimental condition was visualized with bar plots.
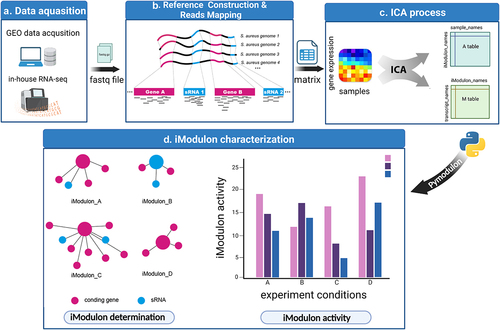
Figure 2. Pangenome-based read mapping methods outperform single-genome-based methods. (a) The cumulative number of conserved genes and total genes with increasing in pangenome size. (b) Number of new genes and unique genes with increasing in pangenome size. (c) Percentage of mapped reads when aligned to pangenomes of different sizes (number of tested RNA-seq datasets = 5). (d) Mapping rates of RNA-seq data derived from 6 S. aureus strains (BPH2947, LAC, MW2, Newman, TCH1516 and BPH2819) when aligned to the pangenome or single genome.
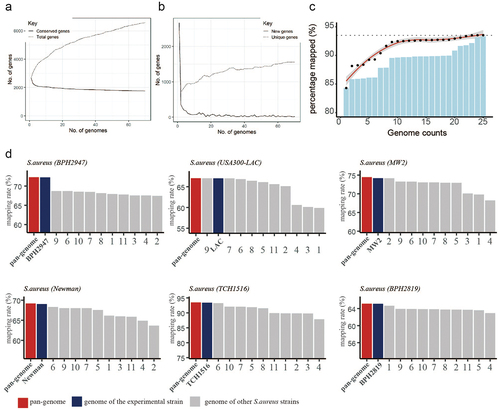
Figure 3. Characteristics of the S. aureus iModulons. (a) Treemap of the 62 S. aureus iModulons. The size of each box represents the explained variance contributed to S. aureus whole transcriptome. Large explained variance indicates more dynamic changes in iModulon activity across conditions. (b) Association between S. aureus iModulons and previously published regulons. iModulons with high fraction of shared genes with published regulon recall were named after the regulon. (c) β-Lactam iModulon components. Each point represents a gene. The x-axis represents the gene ID, and the y-axis indicates the weight of the gene in iModulon. The genes outside the dashed black lines (threshold) are components of iModulon. (d) Bar plot of the iModulon activities for the β-lactam iModulon under different experimental conditions.
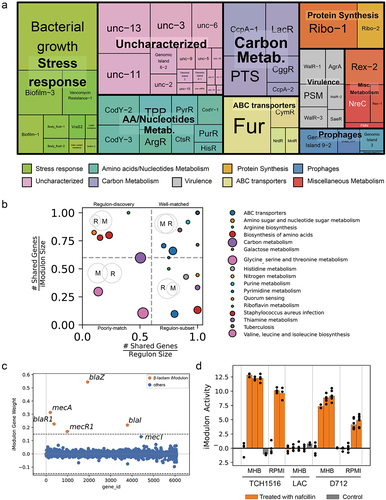
Figure 4. Characteristics of Sau-41. (a) the weight of genes in AgrA iModulon. Genes with prior evidence of Agr regulation are labelled with orange.The genes above the threshold were enriched in AgrA iModulon (b) Distribution of AgrA iModulon components in the MW2 genome. (c) Genomic localization of Sau-41, psmα1–4, and the transcriptional start site (indicated with red arrows) in TCH1516 and MW2 strain. Sau-41 was located within psmα coding sequence and shares a terminator with psmα. TSSs were named after their location in the genome. The TSSs information was derived from previous study [Citation25,Citation26]. (d) Transcription of Sau-41 and RNAIII in USA300 and its isogenic mutant strain ΔagrA at different time points during growth was detected using Northern blot. (e) Quantified transcript level for Sau-41 at various time points was related to RNAIII. The transcript level of each RNA at the first time point was normalized to 100%.
![Figure 4. Characteristics of Sau-41. (a) the weight of genes in AgrA iModulon. Genes with prior evidence of Agr regulation are labelled with orange.The genes above the threshold were enriched in AgrA iModulon (b) Distribution of AgrA iModulon components in the MW2 genome. (c) Genomic localization of Sau-41, psmα1–4, and the transcriptional start site (indicated with red arrows) in TCH1516 and MW2 strain. Sau-41 was located within psmα coding sequence and shares a terminator with psmα. TSSs were named after their location in the genome. The TSSs information was derived from previous study [Citation25,Citation26]. (d) Transcription of Sau-41 and RNAIII in USA300 and its isogenic mutant strain ΔagrA at different time points during growth was detected using Northern blot. (e) Quantified transcript level for Sau-41 at various time points was related to RNAIII. The transcript level of each RNA at the first time point was normalized to 100%.](/cms/asset/8a4ed921-a73b-45bf-9edf-0c2dd6a3de20/kvir_a_2228657_f0004_oc.jpg)
Figure 5. Sau-41 represses S. aureus haemolytic activity. (a) Schematic illustration of the mutation of the PSMα4 start codon. (b) Hemolytic ring of the RN4220 strain carrying Sau-41/Sau-41mt overexpression, knockdown or empty vector plasmid grown on sheep blood agar overnight. (c) Hemolytic activity of the RN4220 strain carrying Sau-41/Sau-41mt overexpression, knockdown or empty vector plasmid determined by incubating bacterial supernatants with 3% human red blood cell. PBS was used as a negative control, and H2O was used as a positive control. (n = 3/group) (d) Transcript levels of hla, hld, psmα, psmβ, hlgA, hlgB, and hlgC were investigated by qPCR in strain RN4220 carrying pRMC2 or pRmc2_sau-41 (n = 3/group). (e) Volcano plot showing the haemolytic factor transcriptional differences between strain RN4220 carrying pRMC2 or pRmc2_sau-41 and their corresponding adjusted p value (q value). (f) the transcriptional activity of hla in strain RN4220 was assessed by analysing cells that harboured plasmids capable of either overexpressing or knocking down Sau-41 (n = 3/group). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Sau-41+: RN4220 carrying the pRmc2_sau-41 plasmid, Sau-41mt+: RN4220 carrying the pRmc2_sau-41mt plasmid, Sau-41kd: RN4220 carrying pSd1_sau-41, wild-type: RN4220 carrying the pRMC2 empty vector.
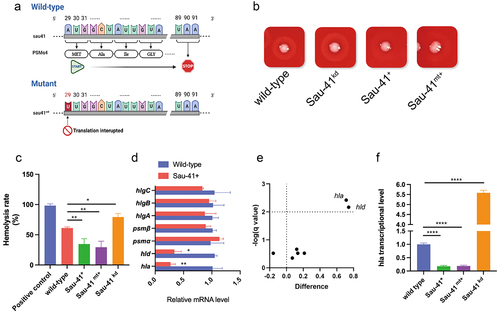
Figure 6. Sau-41 interacts directly with RNAIII. (a) the base pairing region of Sau-41 and RNAIII was predicted with IntaRNA software. (b) Complex formations between RNAIII and Sau-41 were analysed by EMSA. Labeled Sau-41 mixed with increasing concentrations of RNAIII and RNAIII-mutant (5, 10, 25 nM and 50 nM; RNAIII-mutant: wild-type RNAIII with H1-H2 region deleted). Blank: Biotin-labelled Sau-41 without RNAIII/RNAIII-mutant addition. (c) Illustration of the speculated biological function of Sau-41. Sau-41 modulates S. aureus haemolytic activity by binding with RNAIII. (d) Competition between Sau-41 and hlaUTR binding with RNAIII was analysed by EMSA. Biotin-labelled hlaUTR interacted with RNAIII, and increasing concentrations of Sau-41 (10, 25, 50 and 125 nM) were added to interrupt the interaction. Ctrl: hlaUTR RNA (6 nM) interacting with RNAIII (6 nM) without the addition of Sau-41.
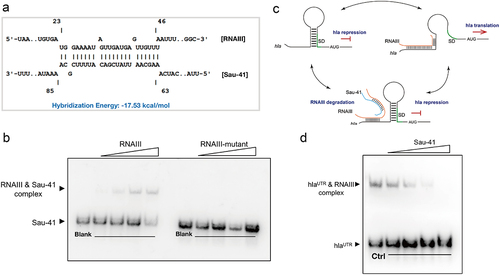
Figure 7. Sau-41 represses S. aureus virulence and alleviates osteolysis in vivo. (a) Illustration of the process of in vivo implant infection model construction. (b) μCT result showing the representative axial, coronal and sagittal views of the implanted femur. (c) 3D reconstruction of μCT data of the infected femur. (d-f) Two hundred sections starting from 30 mm proximal to the femur condyles were quantified for cortical (d) and trabecular (e) bone mineral density and bone volume/total volume (B.V./T.V., F) (n = 6 per group). (g-h): Quantification of the bacterial burden in peri-implant soft tissue (g) and femur (h) (n = 10 per group). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Sau-41+: RN4220 carrying the pRmc2_sau-41 plasmid, Sau-41kd: RN4220 carrying pSd1_sau-41, wild type: RN4220 carrying the pRMC2 empty vector.
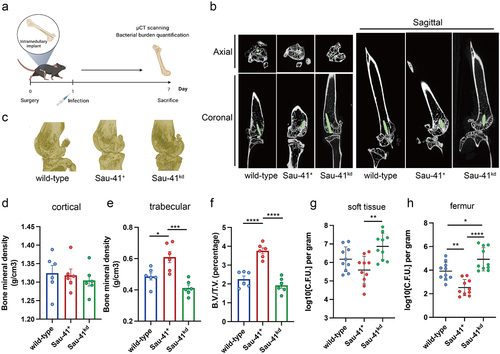
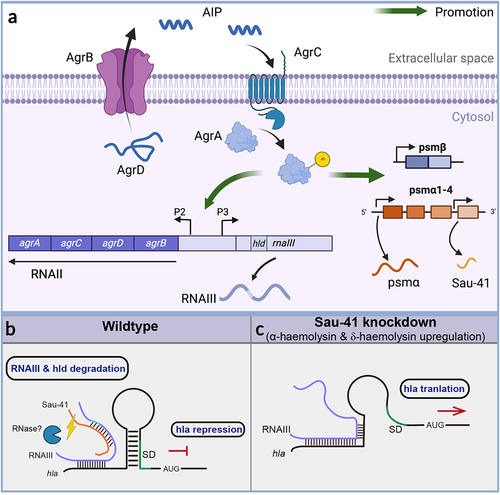
Figure 8 (a) Schematic of the S. aureus Agr system. The AgrD precursor undergoes maturation and export through AgrB, generating the AIP signal. Once the AIP concentration reaches a specific threshold, it triggers the activation of the AgrC-AgrA two-component system. Phosphorylated AgrA then initiates transcription from the P2 promoter, leading to the regulation of auto-feedback. In addition, AgrA facilitates the transcription of RNAIII via the P3 promoter. Furthermore, AgrA enhances the transcription of the psmα and psmβ operons, which encode PSM peptides. Sau-41 is transcribed from the psmα operon. (b-c) the mechanism of Sau-41 in regulating S. aureus virulence. (b) Wild-type: Sau-41 binds with RNAIII thus repressing the expression of α-haemolysin(hla). The Sau-41 and RNAIII complex was probably degraded by RNase III, thus down-regulating the expression of δ-toxin (hld). (c) When Sau-41 was knockdown, RNAIII was preserved and could interact with hla 5’UTR. hla expression was upregulated because its SD sequence was available for ribosome binding.