Figures & data
Table 1. The reagents and antibodies used in this study.
Table 2. Primers used in this study.
Figure 1. HIF-pathway is activated by ISKNV infection. (a) Cells were co-transfected with pGL4-HREs-luc and pRT-TK plasmids and then infected with ISKNV at 6 h post-transfection. The activation of the HIF-pathway was detected using a dual-reporter assay at 48 h post-infection (p.I.). (b,i–l) Spleen samples were collected from infected mandarin fish at 0-, 2-, 4-, and 6-day p.I. (c,e–h) ISKNV-infected cells were collected at 12-, 24-, 48-, and 72-h p.I. Anti-HIF-1α and anti-β-actin (as an internal control) were used in western blot experiments, and the relative expression levels of isknv mcp, glut-1, vegf, and ldha were determined using RT-qPCR. (d) Expression levels of hif-1α after ISKNV infection. Spleen samples were collected from infected mandarin fish at 0-, 2-, 4-, and 6-days p.I. The relative expression levels of hif-1α, glut-1, vegf, and ldha were detected using RT-qPCR.
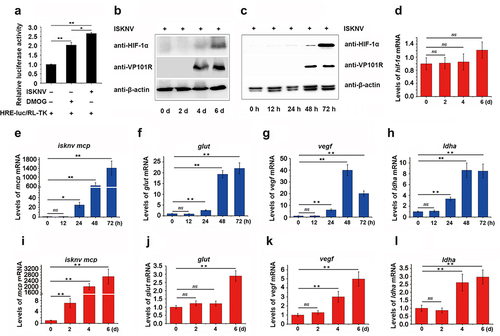
Figure 2. The ISKNV VP077R promotes the HIF-pathway. (a) Analysis of the ankyrin-repeat-containing domain of ISKNV VP077R, VP102R, VP119L, and VP124L. (b–c) The activation of the HIF-pathway was detected using dual-reporter assay at 48 h post transfection. (d–e) The protein levels of HIF-1α in cells were measured after overexpression of pCMV-myc-VP077R or pCMV-myc (control) for 24 h. Cells were treated with DMSO or DMOG before collection and extraction of nuclei. HIF-1α and histone (as an internal control for nuclear protein) were detected using anti-HIF-1α and anti-histone. VP077R and β-actin (as an internal control) were detected using anti-myc and anti-β-actin antibodies. (f–h) The relative expression levels of glut-1, vegf, and ldha were measured in MFF-1 cells transfected with pCMV-myc-VP077R or pCMV-myc (control).
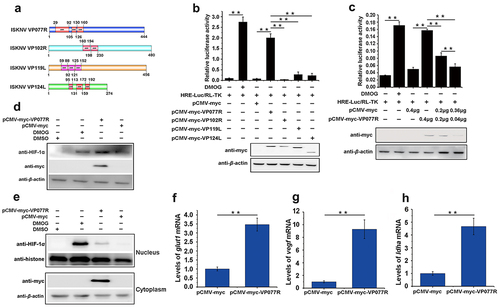
Figure 3. The ISKNV VP077R interacts with VHL and competitively inhibits the interaction between HIF-1α and VHL. (a) Description of VP077R, VHL, and various mutants used in this study. (b) The interaction between VP077R with VHL was detected using CO-IP assays. Cells were co-expressed with myc-VP077R/ha-VHL, myc-VP077R/ha-tag, and myc-tag/ha-VHL. Immunoprecipitation of myc-VP077R was performed using anti-myc antibody, and immunoprecipitation of ha-VHL was performed using anti-ha antibody. (c) Cells were co-expressed with the indicated plasmids. Immunoprecipitation of myc-VP077R was performed using anti-myc antibody, and while immunoprecipitation of the ha-VHL-ΔHIF binding region was performed using anti-ha. (d) Cells were expressed with 6 μg, 3 μg, or 0.6 μg of myc-VP077R, as well as co-expressed with ha-VHL, and flag-HIF-1α. Immunoprecipitation of ha-VHL was performed using anti-ha and detection of flag-HIF-1α was performed using anti-flag. (e) Cells were treated with CHX at 24 h after transfection with the indicated plasmid and then were collected and detected at 0 h, 2 h, 4 h, 6 h, using the anti-flag, anti-myc, anti-ha and anti-β-actin antibodies.
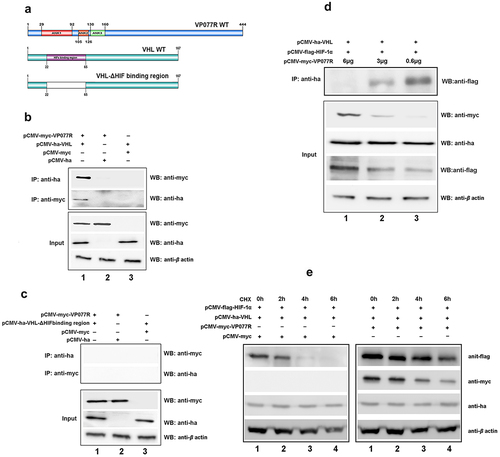
Figure 4. ISKNV VP077R interacts with FIH and promots the degradation of FIH protein by ubiquitin. (a) Description of VP077R, FIH, and various mutants used in this study. (b) The interaction between VP077R and FIH was determined using CO-IP assays. Cells were co-expressed with myc-VP077R/ha-FIH, myc-VP077R/ha-tag, and myc-tag/ha-FIH. Immunoprecipitation of myc-VP077R was performed using anti-myc and immunoprecipitation of ha-FIH was performed using the anti-ha. Used anti-myc, anti-ha, anti-β-actin to detect VP077R, FIH, and β-actin (as an internal control). (c) Cells were co-expressed with ha-FIH and myc-tagged VP077R or its mutants. Immunoprecipitation of myc-tagged VP077R or its mutants was performed using anti-myc, and in turn immunoprecipitation of the ha-FIH protein was performed using anti-ha. (d–f) Cells were co-expressed with myc-VP077R/ha-tagged FIH mutants (lane 1), myc-VP077R/ha-tag (lane 2), and ha-tagged FIH mutants/myc-tag (lane 3). Immunoprecipitation of myc-VP077R was performed using anti-myc, and immunoprecipitation of ha-tagged FIH mutants was performed using anti-ha. (g–h) Cells were treated with CHX and MG132 (right panel)/DMSO (left panel) at 24 h post-transfection with pCMV-myc-VP077R and pCMV-ha-FIH, and then the cells were collected at indicated times. (i) Cells were co-expressed with myc-FIH, ha-ubiquitin, and GFP (lane 1) or GFP-VP077R (lane 2). Immunoprecipitation of myc-FIH was performed using anti-myc and detection of poly-ubiquitin chains was performed using anti-ha. (j) Cells were co-expressed with GFP-VP077R, myc-FIH, and ha-ubiquitin or ha-ubiquitin K48R. Immunoprecipitation of myc-FIH was performed using anti-myc and detection of poly-ubiquitin chains was performed using anti-ha antibody.
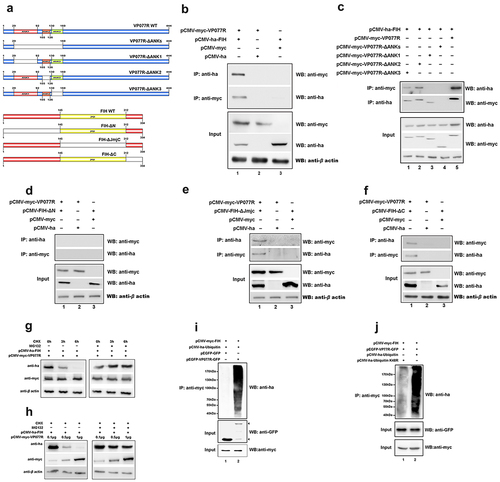
Figure 5. The “FIH – ISKNV VP077R – Skp1 complex” promotes FIH ubiquitination and degradation. (a) Cells were co-expressed with myc-VP077R/flag-Skp1, myc-VP077R/flag-tag, or myc-tag/flag-Skp1. Immunoprecipitation of myc-VP077R was performed using anti-myc, and immunoprecipitation of flag-Skp1 was performed using anti-flag. (b) Cells were co-expressed with ha-FIH/flag-Skp1, ha-tag/flag-Skp1, or ha-FIH/flag-tag. Immunoprecipitation of ha-FIH was performed using anti-ha, and in turn immunoprecipitation of flag-Skp1 was performed using anti-flag. (c) Cells were co-expressed with myc-VP077R or myc-tag, ha-FIH, and flag-Skp1. Immunoprecipitation of flag-tagged Skp1 protein was performed using anti-flag. (d–e) Cells were treated with CHX at 24 h after transfection with the indicated plasmid. Detections of VP077R, FIH, Skp1, and β-actin (as an internal control) were performed using anti-myc, anti-ha, anti-flag, and anti-β-actin, respectively.
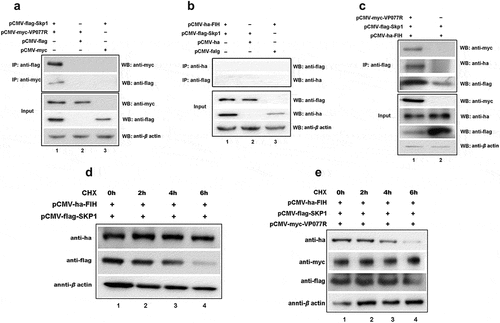
Figure 6. The activation of the HIF-pathway promotes ISKNV replication. (a–f) Cells were treated with DMOG (1 mM) or BAY87–2243 (100 nM) at 4 h after ISKNV infection, and then the levels of ISKNV infection were determined at indicated times. (a,d) levels of isknv mcp mRNA were detected using RT-qPCR. (b,e) Levels of viral genomic DNA were detected using absolute qPCR. (c,f) Viral titres (TCID50) of the cell lysates were determined by repeated freezing and thawing three times. (g–n) Fish were injected intraperitoneally with DMOG or BAY87–2243 at 4 h after ISKNV infection, and were given supplementary injections every three days. The spleen samples were separated and then the viral loads (g,k) and the transcription levels of the downstream genes (glut-1, vegf, and ldha) of the HIF-pathway (h–j, l–n) were determined.
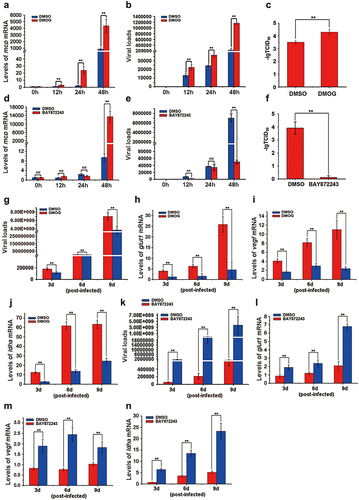
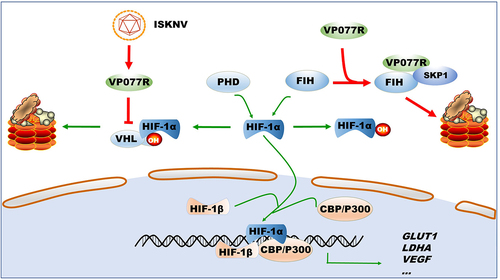
Figure 7. Model of how ISKNV protein VP077R regulates the HIF-pathway. ISKNV VP077R binds to VHL and competitively inhibits its interaction with HIF-1α, stabilizing the level of HIF-1α protein. Additionally, ISKNV VP077R interacts with FIH, promoting its ubiquitin-dependent degradation and maintaining the transcription activity of HIF-1α.
Data Availability statement
The data that support the findings of this study are openly available in figshare at https://figshare.com/, reference number [10.6084/m9.figshare.25565820].
