Figures & data
Table 1. All NCBI open reading frame (ORF) finder sequence matches for Tpl94D in the original 12 sequenced Drosophila species
Table 2. Best NCBI nucleotide BLAST sequence matches and orthologs for Tpl94D (GI: 24649165)
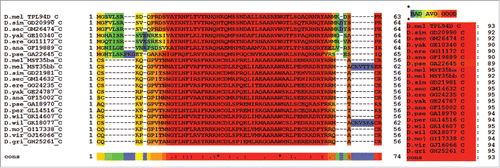
Figure 1. T-Coffee alignment of Tpl94D for melanogaster species subgroup, D. ananassae, and D. pseudoobscura. T-Coffee conserved region alignment for Tpl94D. Key on the bottom right shows 87 consensus score for all sequence matches.
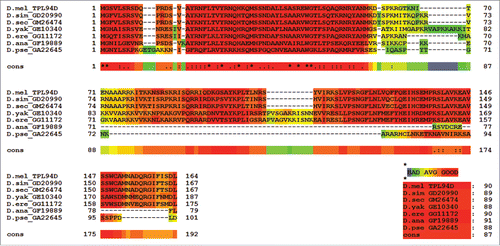
Figure 2. CLUSTAL Omega Alignment of Tpl94D sequence matches CLUSTAL Omega alignment of the best Tpl94D sequence matches in the sequenced 12 Drosophila species.

Table 3. NCBI protein BLAST Tpl94D (GI: 24649166) orthologs
Table 4. Amino acid analysis for Tpl94D orthologs
Figure 3. CLUSTAL Omega Alignment of Tpl94D with mammalian TP2 CLUSTAL Omega alignment of the orthologs for Tpl94D and transition protein 2 from M. musculus, R. norvegicus, B. taurus, and H. sapiens.

Figure 4. Phyre2 Tpl94D best sequence matches conserved DNA binding region Tertiary structure alignment of a wire frame model for the Tpl94D orthologs. The different colors indicate each of the species indicated on the bottom right.
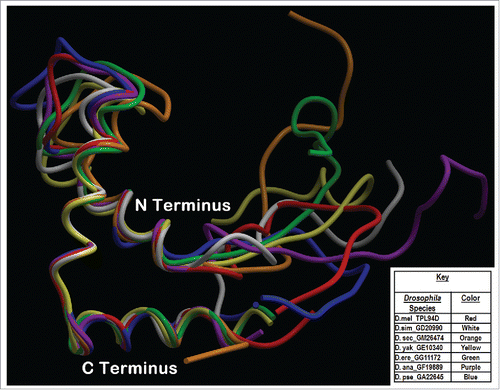
Table 5. Detailed analysis of conserved functional groups found in Tpl94D orthologs
Table 6. Detailed analysis of functional groups found in Tpl94D whole protein sequence matches
Table 7. Ovaries vs. testes transcriptome Cuffdiff 2.0.2 RNA-Seq analysis summary
Figure 5. Phyre2 Tpl94D orthologs, MST35Ba, MST35Bb orthologs, Dpse GA18970 Exon 1 (GA31252) best sequence matches conserved DNA binding region Tertiary structure alignment of a wire frame model for the Tpl94D orthologs. The different colors indicate each of the species shown on the bottom right.
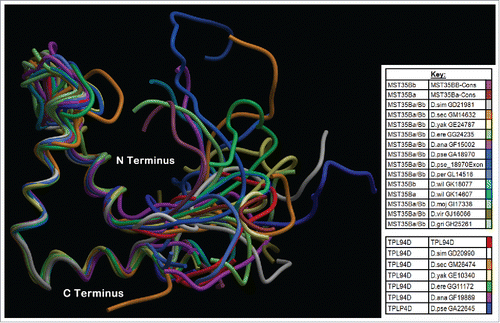
Figure 6. T-Coffee Alignment of Tpl94D orthologs with MST35Ba and MS35Bb orthologs. A T-Coffee alignment of the whole protein Tpl94D conserved region and the whole proteins of the Drosophila protamine-like proteins found in the 12 sequenced Drosophila species shows high conservation of the conserved regions with a T-Coffee consensus score of 72. D. pseudoobscura GA18970s first exon (GA31252) was used due to size length of the whole protein being 569 amino acids.
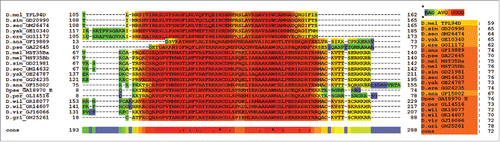
Figure 7. T-Coffee Alignment of Tpl94D and Drosophila protamine-like protein (MST35Ba and MST35Bb) conserved regions. T-Coffee alignment of the DNA binding-HMG box conserved regions in Drosophila Tpl94D and Drosophila protamine-like proteins (MS35Ba and MST35Bb) orthologs. The area in red indicates strong conservation. Consensus score equals 93. Also D. pseudoobscura GA18970s first exon (GA31252) contains the conserved region for the D. pseudoobscura MST35Ba/Bb ortholog.