Figures & data
Figure 1 Flow chart showing the process for the Mendelian randomization analyses. eQTL: expression quantitative trait loci; ALS: amyotrophic lateral sclerosis; MAF: minor allele frequency; GWAS: genome-wide association studies; SMR: summary data-based mendelian randomization; IVW: inverse-variance weighted.
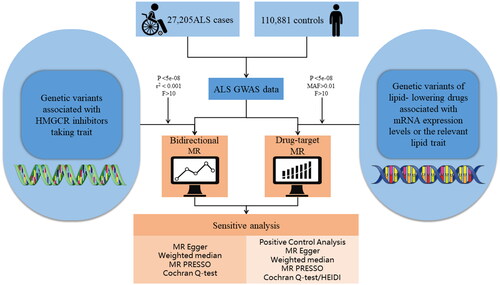
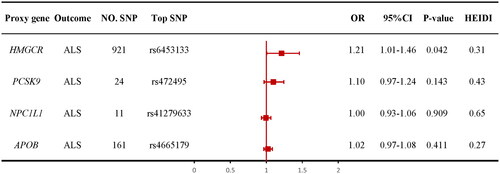
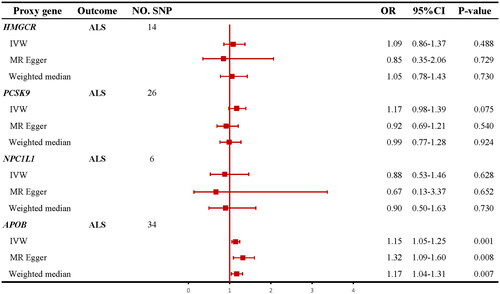
Figure 2 Information of genetic instruments. HMGCR: HMG-CoA reductase; PCSK9: proprotein convertase subtilisin/kexin type 9; NPC1L1: Niemann–Pick C1-like 1; APOB: apolipoprotein B; SMR: summary data-based mendelian randomization; eQTL: expression quantitative trait loci; MAF: minor allele frequency; SNP: single nucleotide polymorphism; LDL: low-density lipoprotein.
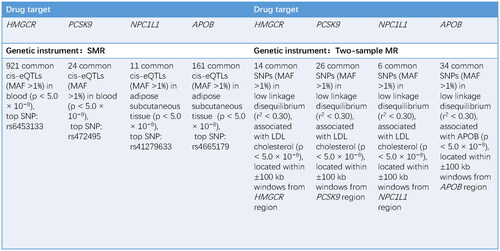
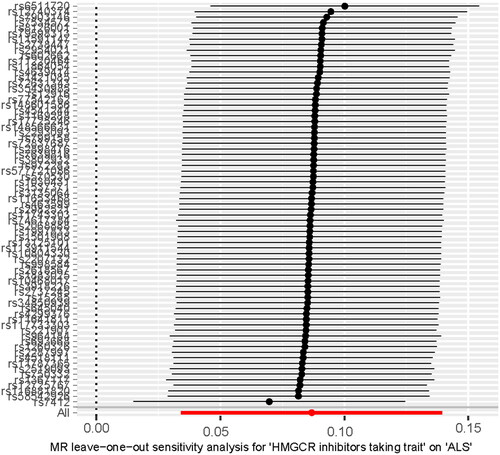
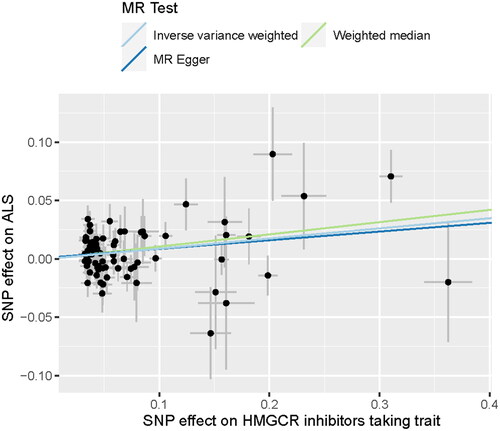
Figure 3 Scatter plot showed the SNP effects on HMGCR inhibitors-taking trait versus ALS. HMGCR: HMG-CoA reductase; ALS: amyotrophic lateral sclerosis.
Supplemental Material
Download Zip (1.2 MB)Data availability statement
The summary statistics used in the current study are available from the corresponding author upon reasonable request. Detailed information on the summarized data sources for the instrumental variables is presented in Supplementary Table S1.