Figures & data
Figure 1. Applications and Opportunities (in blue) for mammalian libraries in antibody engineering and hit discovery. Libraries from precious diversities such as elite responders can be cloned in desired therapeutic formats and produced in display or secretion mode for repeated interrogation by flow cytometry or microfluidics, respectively. Hit candidate recovery by PCR gene amplification & transient re-production or single cell sub-cultivation enable secondary hit confirmation. Mammalian libraries offer several advantages for each step highlighted in blue and discussed in the main text.
Figure 2. Differentiating display levels of exemplary Bococizumab versus Cetuximab and sorting for display and target specificity. (a)-(d) Spiking of cells displaying anti-IgG1 stained Cetuximab (gate C) and Bococizumab (gate B) in ascending ratios of 1:10,000 to 1:10. Labels represent percentage from pre-gated populations (Figure S5(a)). Increasing prevalence of events in gate C match the higher Cetuximab to Bococizumab ratio. 1:10 spiked cells were stained with either anti-IgG (d) or target antigen EGFR and PCSK9, respectively (h). Percentage of events in gates B and C equals with prevalence of specifically target-binding cells in (h) Q1 and Q3, respectively, indicating clone-dependent display levels observed in (a)-(d). A 1:10,000 mixture was sorted for highest display and subsequently for EGFR specificity (please refer to Figure S4 for full gating strategy and sorting plots). (e) An analytical FACS post display sorting indicates enrichment for EGFR-positive Cetuximab displaying cells. Subsequent sorting for EGFR specificity yields highly enriched Cetuximab displaying cells detected via EGFR binding (f) or anti-IgG1 staining (g). (h) A 1:10 clone mixture generated with optimized conditions when stained for EGFR or PCSK9 binding yields ∼38% monoclonal versus ∼1.4% oligoclonal cells, hence ∼96% monoclonality in displaying cells.
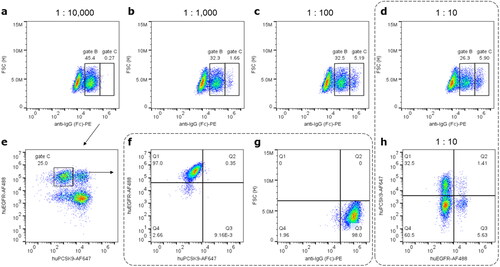
Figure 3. Secretion, cell viability within and PCR recovery rates post microfluidic screening. (a) Bulk measured specific productivity of cells seeded at 0.2x106 vc/ml and 0.5x106 vc/ml, respectively. (b) Exemplary image of CHO cell secretion in pen on Beacon device and accumulated percentage of cells with confirmed secretion over time (c). (d) Cell growth in 8 representative pens over three days, indicating potential advantages over primary cells for longer-lasting functional screens. (e) Full mAb PCR recovery rates in relation to time on Cyto-Mine device. In comparison to primary cells after prolonged incubation (green and purple), CHO cells (blue circles) yield high recovery rates similar to hybridoma (red squares), indicating high retained viabilities and an overall robust screening system.
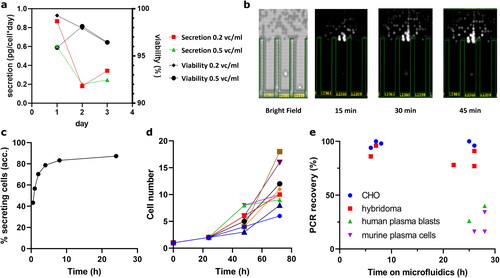
Figure 4. Therapeutic and complex formats can be produced in CHO library systems. (a) A stably integrated, secreted IgG1-VHH fusion targeting TMA and an TNFR showed simultaneous antigen binding in supernatant after bulk cultivation. 1, TMA loading; 2, quenching and baseline; 3, IgG-VHH binding from SN; 4, TNFR binding. (b) Similarly expressed, a tetra-specific VHH-SEED fusion bound all four targets, indicating the applicability of the system for complex formats including heterodimeric Fc fusions. Aligned to step 1. 1, loading of SEED SN; 2, baseline; 3, NKG2D; 4, IL-6R; 5, EGFR; 6, Her2.
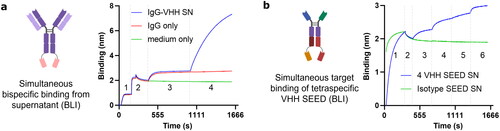
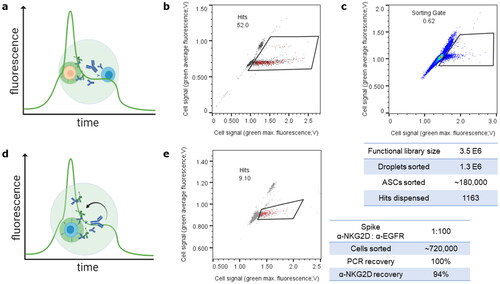
Figure 5. High throughput screening for binding to co-encapsulated target cells or in autocrine setup with increased sorting throughput. (a) Scheme representing a droplet and setup for sorting of cellular binding antibodies secreted from CHO cells (blue) to co-encapsulated target cells (orange) and detected via anti-IgG-488, and overlaid plot indicating a peak in cell-targeted fluorescence allowing sorting for green peak versus average fluorescence signals. (b) Dispensing plot of positive run with Cetuximab IgG1 secreting cells spiked 1:100 in non-secreting cells, co-encapsulated with EGFR-positive A431 cells and successfully sorted for cellular binding. (c) Sorting plot and statistics of interrogation of a cLC diversity against a tumour-specific mucin antigen via cellular binding on HeLa adenocarcinoma cells. Dispensed hits were subjected to Next Generation Sequencing. (d) Scheme of sorting in an autocrine setup. The CHO cell (blue) is both recombinantly expressing the antigen of interest and secreting one antibody candidate. Sorting is feasible for green peak fluorescence as in (a). (e) Dispensing plot and statistics for proof-of-concept autocrine cellular binding exemplarily to NKG2D by targeted anti-NKG2D or irrelevant anti-EGFR VHH-Fc binders applied in a 1:100 ratio. Note both a comparatively high throughput and high recovery rates indicating a robust screening system. For full sorting and dispensing plus gating information refer to Figure S5.
Supplemental Material
Download MS Word (1.6 MB)Data availability statement
The data that support the findings of this study are available from the corresponding author, AD, upon reasonable request
