Figures & data
Figure 1. Sequence comparisons between SARS-CoV-2, the original GX_P2V sample and the pangolin coronavirus GX_P2V isolate. The GX_P2V isolate (GX_P2V(short_3UTR)) is a variant with two interesting mutations: one nonsynonymous mutation (C to U) in the final amino acid codon of the nucleoprotein and one 104-nt deletion in the 3'-UTR. (A) Alignment of the 3'-terminus sequences of the nucleoprotein genes. (B) Alignment of the partial 3'-UTR sequences. Dots represent residues that are identical to those in SARS-CoV-2 and hyphens represent nucleotide deletions.

Figure 2. Growth of the pangolin coronavirus GX_P2V variant in two non-human primate cells (BGM and Vero) and human Calu-3 cells. (A) BGM and Vero cells infected with GX_P2V(short_3UTR) show similar cytopathic effects: limited cell degeneration at 24 h postinfection, and more evident cell degeneration and cell rounding after 48 h postinfection. Calu-3 cells infected with GX_P2V(short_3UTR) showed no cytopathic effects. Scale bars, 100 μm. (B) The replication of GX_P2V(short_3UTR) in BGM and Calu-3 cells at different time points was detected by immunofluorescence assays with anti-sera from GX_P2V(short_3UTR)-infected hamsters. Nuclei were stained with DAPI (blue). Scale bars, 50 μm. (C) GX_P2V(short_3UTR) forms small plaques in BGM and Vero cells at five days postinfection, while no plaque was produced in Calu-3 cells. The small white dots represent plaques. (D) The diameter of plaques (blue circle) was measured by using Image Pro plus 6.0 software. GX_P2V(short_3UTR) forms bigger plaques in Vero than in BGM (p < 0.0001).
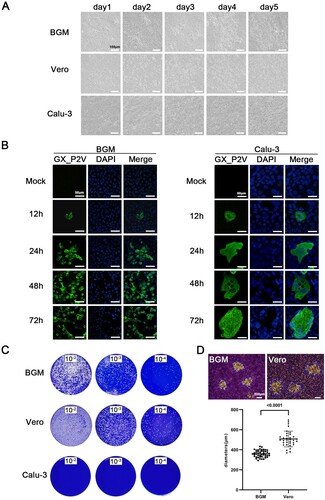
Figure 3. Replication kinetics of GX_P2V(short_3UTR) in BGM, Vero and Calu-3 cells. Results are expressed as the mean of three biological replicates. (A) Viral RNA copies were determined by qRT-PCR. (B) Viral titres were titrated by using a standard TCID50 assay. Compared to BGM and Calu-3, Vero cells appear to produce different viral titres at 24 and 120 h postinfection (***p < 0.001).
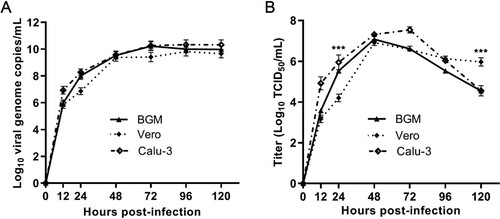
Figure 4. Intranasal infection of golden hamsters shows GX_P2V(short_3UTR) had a short duration of viral replication and caused no gross pathology in the respiratory tracts. (A) Scheme of intranasal infection. (B) Both mock and GX_P2V(short_3UTR)-infected hamsters had normal body weight increases and no apparent clinical symptoms in the two-week observational period. Individual data points represent percent weight changes compared to day zero, or cumulative clinical score. (C) Representative images of lungs and tracheas from mock hamsters, 1 × 104, and 1 × 105 TCID50 infected hamsters at indicated time points. All organs had no gross pathology. (D-I) Viral RNA loads (D-F) and viral titres (G-I) in homogenate supernatants of lungs (D, G), tracheas (E, H) and turbinates (F, I) of GX_P2V(short_3UTR)-infected hamsters at different days post-inoculation were determined by qRT-PCR and the TCID50 assay, respectively. The detection limit is shown by the dotted line. Error bars represent means ± SD.
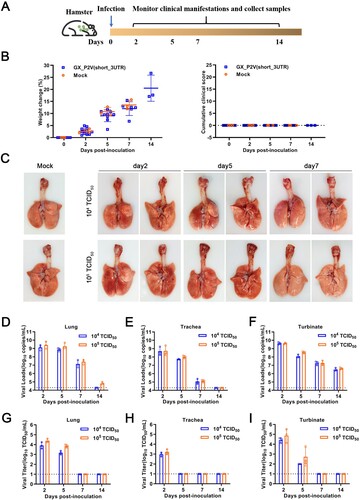
Figure 5. Histopathological analyses of lung and trachea tissues from intranasally infected hamsters. H&E 400 × are the magnifications of the region in the corresponding red box in H&E 100 × . (A) 1 × 104 TCID50 infected group (n = 3), no pathological changes were observed in the lungs. (B) 1 × 105 TCID50 infected group (n = 3), lungs had no tissue lesions. Few inflammatory cells of lymphocytes and neutrophils associated with blood vessels were observed at five days postinfection but not at other time points. (C) Representative images show that tracheas of both infection dose groups had no significant histopathological changes at all time points post-infection.
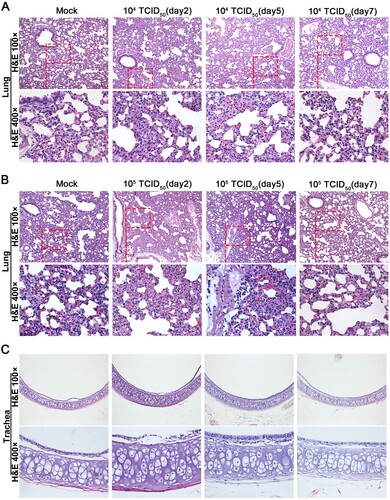
Figure 6. Intragastrical inoculation of GX_P2V(short_3UTR) in golden hamsters established limited infection in the respiratory tract. (A) Viral RNAs were detected in samples of tongues and feces from the above golden hamsters with intranasal infection, suggesting established infections in the gastrointestinal tract. (B) Scheme of intragastrical infection. Samples of tongues and feces were collected at indicated time points. (C) Body weights of all golden hamsters had continuous increases. (D) No clinical symptoms of infection were identified. (E) Limited viral RNAs in the lungs, but not in other organ tissues were detected by qPCR. (F) No histopathological changes were identified in lung tissues of mock and GX_P2V(short_3UTR)-inoculated golden hamsters. (G) Viruses were not detected in feces from golden hamsters following intranasal or intragastrical inoculation.
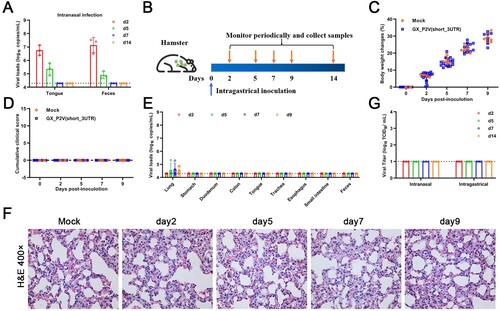
Figure 7. Intranasal inoculation of GX_P2V(short_3UTR) in both young and aged BALB/c mice. (A) Scheme of intranasal infection. Young and aged BALB/c mice were intranasally infected with mock or 1 × 105 TCID50 of GX_P2V(short_3UTR). Various infection outcomes and tissue samples were collected at indicated time points. (B) Young mice had constant body weight increases after infection. (C) Intranasal inoculation of GX_P2V(short_3UTR) in young and aged BALB/c mice produce no detectable gross pathology. (D) Viral RNA loads in infected lungs of both young and aged BALB/c mice were determined by qPCR. (E-F) Representative H&E images of infected lungs from young (E) and aged (F) BALB/c mice. (G) No viral titres were detected in infected lungs from both young and aged BALB/c mice.
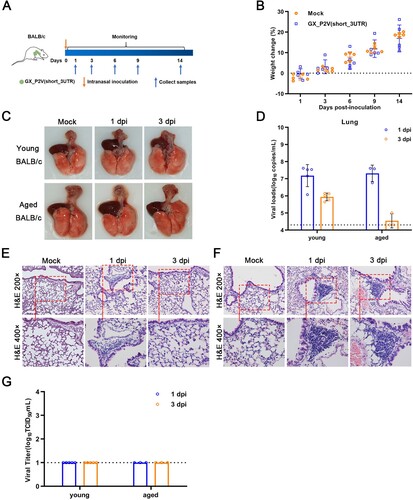
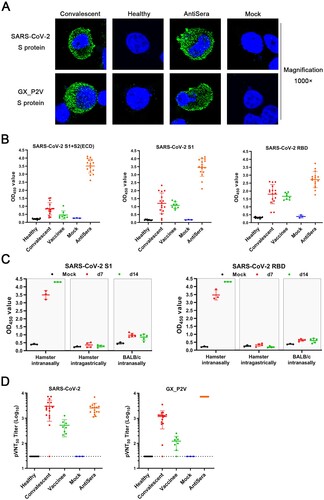
Figure 8. Serological cross-reactivity and cross-neutralization between GX_P2V and SARS-CoV-2. (A) Immunofluorescence assays with 293T cell expressing spike proteins (green) of SARS-CoV-2 and GX_P2V, using sera from convalescent COVID patients, healthy donors, and GX_P2V(short_3UTR)-infected golden hamsters. Nuclei were stained with DAPI (blue). (B) Three commercial ELISA kits for detecting antibodies against SARS-CoV-2 S1 + S2 (ECD), S1, and RBD were used for measuring antibodies in sera from healthy donors (n = 10), convalescent COVID patients (n = 16), vaccinees of inactivated COVID-19 vaccines (n = 8), mock-infected golden hamsters (n = 3), and GX_P2V(short_3UTR)-infected golden hamsters (n = 16), respectively. (C) Cross-reacting antibodies against SARS-CoV-2 S1 and RBD in sera from GX_P2V(short_3UTR)-infected golden hamsters and BALB/c mice were measured by using the aforementioned ELISA kits. No cross-reacting antibodies against SARS-CoV-2 S1 and RBD were detected in sera from intragastrically infected golden hamsters or intranasally infected BALB/c mice. (D) Neutralizing antibodies against two pseudoviruses, SARS-CoV-2 and GX_P2V, were detected in sera as described in (B). The error bars indicate means ± SD.