Figures & data
Figure 1. The location and type of collection site of the samples with whole genome sequences. Samples with whole genome sequences were collected in Amibara (n = 11), Gewane (n = 3), and Chifra (n = 3) from Afar region, Babile (n = 3) from Oromia region, and Akaki (n = 5) in the capital city, Addis Ababa.
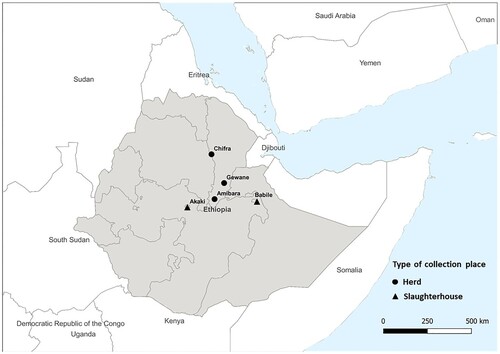
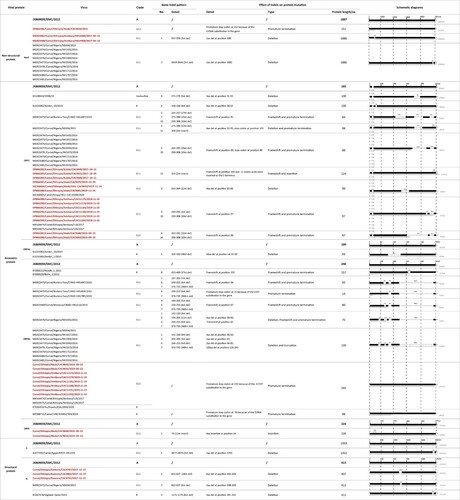
Figure 2. (A) Phylogenetic analysis of MERS-CoV whole genome sequences obtained in this study using IQ-TREE. The tree is rooted against MERS-CoV-related bat coronavirus Coronavirus Neoromicia/PML-PHE1/RSA/2011(GenBank: KC869678.4), which was removed from the tree due to the long branch length. Selective bootstrap values are shown. The MERS-CoV clades designations are denoted based on the previous studies [Citation28,Citation47]. Whole genome sequences labelled in red are from this study and those in black are sequences downloaded from Genbank. S gene partial sequences from the previous study with the length of 1046 bp are labelled with asterisk [Citation28]. Indel patterns of nsp3, ORF3, ORF4a, ORF4b, ORF5, S, and N genes observed in the sequences are shown in the table within the tree (S gene partial sequences and sequence MK967708/Egypt/Camel/AHRI−FAO−1/2018 are excluded). Two clade B Saudi Arabian viruses that show deletions in ORF4b are included for comparison. The sampling places (Amibara, Chifra, Gewane, Babile, Akaki) of the sequences are labelled in the tree. (B) Details of the indel patterns of the sequences in the phylogenetic trees.
![Figure 2. (A) Phylogenetic analysis of MERS-CoV whole genome sequences obtained in this study using IQ-TREE. The tree is rooted against MERS-CoV-related bat coronavirus Coronavirus Neoromicia/PML-PHE1/RSA/2011(GenBank: KC869678.4), which was removed from the tree due to the long branch length. Selective bootstrap values are shown. The MERS-CoV clades designations are denoted based on the previous studies [Citation28,Citation47]. Whole genome sequences labelled in red are from this study and those in black are sequences downloaded from Genbank. S gene partial sequences from the previous study with the length of 1046 bp are labelled with asterisk [Citation28]. Indel patterns of nsp3, ORF3, ORF4a, ORF4b, ORF5, S, and N genes observed in the sequences are shown in the table within the tree (S gene partial sequences and sequence MK967708/Egypt/Camel/AHRI−FAO−1/2018 are excluded). Two clade B Saudi Arabian viruses that show deletions in ORF4b are included for comparison. The sampling places (Amibara, Chifra, Gewane, Babile, Akaki) of the sequences are labelled in the tree. (B) Details of the indel patterns of the sequences in the phylogenetic trees.](/cms/asset/cd8e8e97-08fe-401c-819d-deb706f0dd88/temi_a_2164218_f0002_oc.jpg)
Figure 3. Schematic of the indels and their impact on encoded proteins. A clade A prototype virus, JX869059/EMC/2012 served as reference.