Figures & data
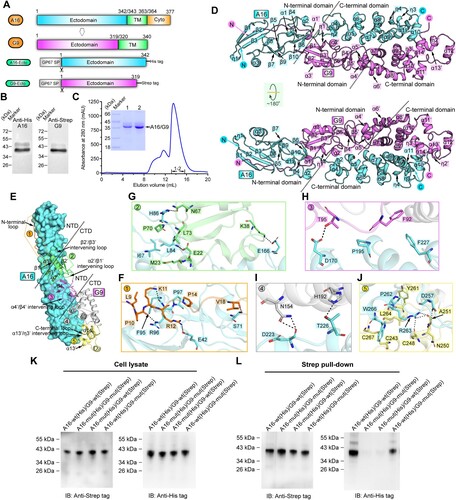
Figure 1. Structure of vaccinia virus A16/G9 sub-complex. (A) A schematic view of the protein-engineering strategy used to yield vaccinia virus A16-Ecto-His/G9-Ecto-Strep sub-complex. The transmembrane domain (TM), the ectodomain, and the cytoplasmic domain (Cyto) are individually marked with the boundary-residue numbers. (B) Identification of A16-Ecto-His/G9-Ecto-Strep hetero-complex using western blot assay. (C) Solution behaviour of A16-Ecto-His/G9-Ecto-Strep subcomplex on a Superdex 200 Increase 10/300 GL column. The inset figure shows the SDS-PAGE analysis of the pooled samples. (D) Overall structure of the hetero-dimer formed between A16 (cyan) and G9 (violet). The secondary structural elements are labelled. Both A16 and G9 could be subdivided into two domains, the boundary of which is highlighted with the dashed lines. (E-J) The atomic binding details between A16 and G9. The binding interface is subdivided into five patches (Patch1-Patch5) based on the G9 components involved in A16-engagement, which is depicted in panel (E). For clarity, only those residues providing important H-bond and hydrophobic interactions are shown and labelled. Dashed lines indicate hydrogen bonds. A full list of the inter-chain contacts, including H-bonds and vdw contacts, is summarized in Supplementary Table S2. (F) Amino acid interaction details in Patch1. (G) Amino acid interaction details in Patch2. (H) Amino acid interaction details in Patch3. (I) Amino acid interaction details in Patch4. (J) Amino acid interaction details in Patch5. (K-L) Verification of the A16/G9 interaction via mutagenesis. His-tagged A16 proteins (wt and mutant) and Strep-tagged G9 proteins (wt and mutant) were co-expressed in cells. G9 was then precipitated using the streptactin resin and the co-precipitated A16 was analysed and compared. (K) Western-blot analyses of the cell lysates. (L) Western-blot analyses of the pulled samples.
Supplemental Material
Download MS Word (3.9 MB)Data availability
The datasets used and/or analysed during this study are available from the corresponding author on reasonable request. Atomic coordinates and structure factors for the reported crystal structure has been deposited into the Protein Data Bank under accession number 8GP6.
