Figures & data
Figure 1. SARS-CoV-2 3CLpro transfection induced cytotoxicity and suppressed reporter expression in 293T cells, and it can be rescued by treatment with the specific inhibitor. (A) Schematic of the BRET, reporters, and 3CLpro expression constructs. (B) The “WT” represents wild-type 3CLpro co-transfected with reporter as indicated, the “C145A” indicates the C145A mutant co-transfected, and the “WT+GC376” is the same as the “WT” but with a treatment of GC376 at 20 μM. The “Mock” is native cells without any treatment. (C) Western blotting results obtained from cells treated as described in (B). The arrows point to mKate2/EGFP (26 kD), 3CLpro (36 kD). Bands at about 55 kD is α-tubulin. (D) Different expression of reporters in cells treated with WT+GC376 vs. WT or C145A. Data were presented as mean ± SD (n = 3), and the significant difference was determined using the Student t-test. (E) The dual-reporter construct was used. Data with n = 2 was analysed using an ANOVA with a post-hoc Tukey's HSD test. *: p < 0.05; **: p < 0.01; ***: p < 0.001; ****: p < 0.0001; ns: not significant.
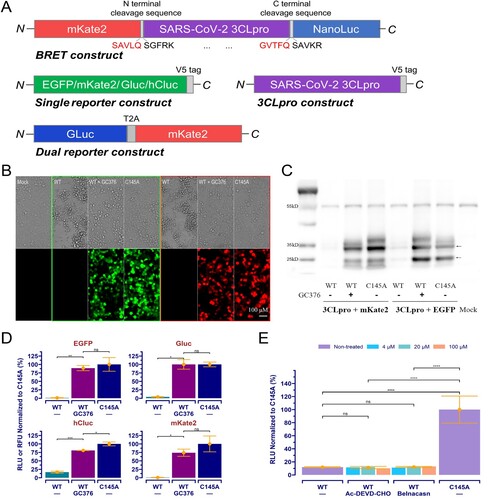
Figure 2. The orthogonal dual reporter assay provides a sensitive and quantitative measurement of 3CLpro activity and inhibition by specific inhibitors in living cells. (A) Correlation analysis of reporter readouts vs 3CLpro activity. The data points plotted are from single experiment (n = 3), and the lines with grey shading (95% CIs) were predicted by fitted linear regression. (B) and (C) Dose-response curve and IC50 of the GC376 and PF-00835231 in the Gluc assay and mKate2 assays, respectively. The date point plotted is from two repeated experiments (n = 4). (D) Ebselen and masitinib exhibited no inhibition on 3CLpro in assay. The Gluc assay data from a single experiment (n = 3) is plotted.
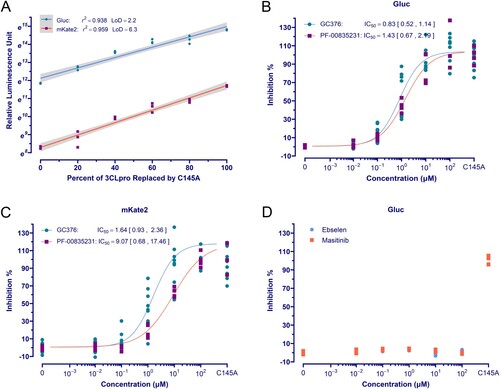
Figure 3. High throughput screening. (A) The schematic diagram of the HTS setting. GC376 for positive control was used at 20 μM, and 0.2% DMSO was used for negative control. (B) Dose-response curve and IC50 of hits using the Gluc assay. The data points plotted are from two repeated experiments (n = 3). (C) and (D) The results of HTS. The red dashed line is the mean value of positive controls, and the blue dashed line indicates the threshold for hit selection. The yellow triangles with annotation are GC376 (2.00 μM) and PF-00835231 (1.25 μM) randomly interpolated for performance validation.
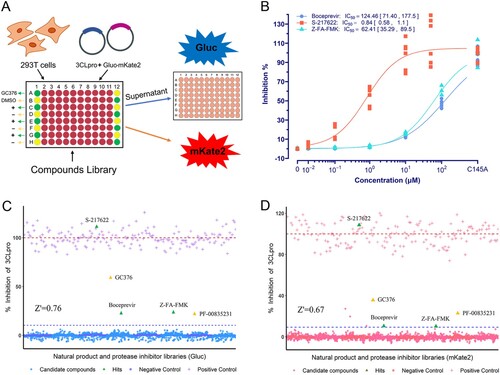
Table 1. 3CLpro mutations investigated in this study.
Figure 4. Effect of mutations on 3CLpro activity and its susceptibility. (A) Activity assay of 3CLpro mutants investigated. The data is presented as the mean with raw data points from three repeated experiments (n = 3). The significant difference compared to WT was determined using the ANOVA with a post-hoc Tukey's HSD test. (B ∼ D) The susceptibility of 3CLpro mutants to GC376, PF-07321332, and S-217622. The data points plotted are from three repeated experiments (n = 3).
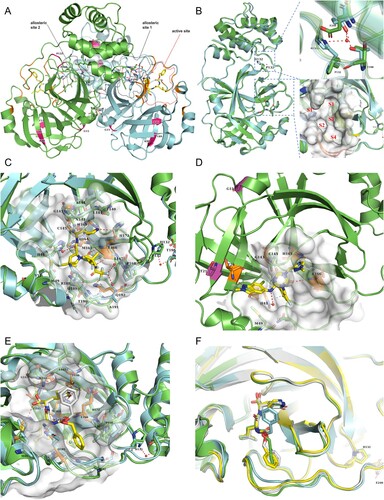
Figure 5. SARS-CoV-2 3CLpro structures with inhibitors bound. (A) 3CLpro homodimer (green and cyan) with GC376 bound (PDB ID 6WTT). The active site and two allosteric sites are circled. Mutation sites in two promoters are indicated in hot pink. Hydrogen bonds at the dimerization interface are also indicated (red dashed lines). (B) Comparison of the GC376-cocrystal structures of P132H (cyan; PDB ID 7TOB) and WT (green). GC376 and residues needed for comparison are shown as sticks. A close-up view of position 132 (top) and the substrate-binding pocket (bottom) is shown in the right panel. Water molecules are shown as red spheres. Dashed red lines indicate hydrogen bonds. S1’ and S1–S4 in red indicate sub-pockets. (C) Close-up view of PF-07321332 binding to 3CLpro (green; PDB ID 7VH8) with superimposition of P132H (cyan). The inhibitor is shown as a yellow stick. Residues participating in binding are shown as sticks, and the hydrogen bond acceptors are coloured orange. Water molecules and hydrogen bonds are shown as described in (B). (D)S-217622 binding to the 3CLpro active site (PBD ID 7VU6). Residues involving mutation are shown as purple sticks, and the π−π stacking is indicated by a dashed cyan line. Inhibitor and residues involving binding are shown as described in (C), except for the hydrogen bond between T21 and T26, which is shown in yellow. (E) Superimposition of GC376 binding to WT (green) and P132H (cyan) 3CLpro. GC376 is shown as yellow sticks or grey for its binding to WT or P132H, and residues involving inhibitor binding are shown as described above. Hydrogen bonds are indicated by red or yellow dashed lines for WT or P132H binding. (F) Superimposition of the GC376-bound P132H mutant (cyan) and WT 3CLpro (green, yellow, and grey for the three protomers in the asymmetric trimeric subunit).
Supplemental Material
Download MS Excel (64.5 KB)Supplemental Material
Download MS Excel (309.7 KB)Supplemental Material
Download MS Word (137.3 KB)Data availability statement
All data to understand and assess the conclusions of this study are available in this published article. The raw data that support the findings of this study are available from the corresponding author upon reasonable request.
