Figures & data
Figure 1. The Pex14 roles in peroxisomal biology of B. bassiana. (a) Sub-cellular localization of BbPex14. BbPEX14 was fused to mCherry gene and transformed into a strain in which the peroxisomes were labeled with green fluorescent protein. The transgenic fungus was cultured under submerged (SDB) and aerial (SDAY) conditions. (b) Translocation of peroxisomal proteins. The fusion proteins GFP-SKL and Thi-GFP represented the proteins with peroxisomal targeting signal (PTS) type 1 and 2, respectively. The translocation of fusion proteins was examined in the wild-type and ΔBbpex14 mutant strains. Fluorescent signals were detected under a laser scanning confocal microscope. Bars: 5 µm.
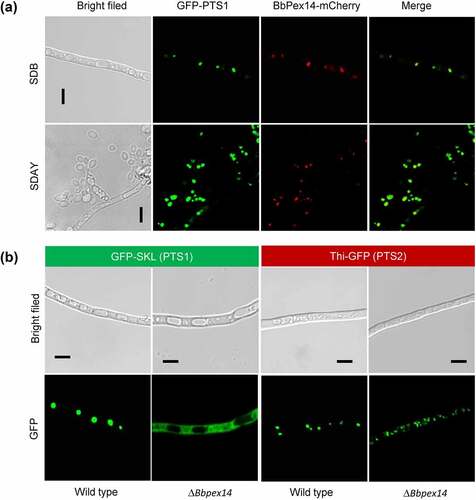
Figure 2. Phenotypic assay for ΔBbpex14 mutant strain. (a) Fungal stress response was determined on CZA plate included with stress chemicals. Colony diameter was examined 7 d post incubation, using CZA as control. Fungal conidiation (b) and blastospore formation (c) were quantified on SDAY plate and SDB broth, respectively. Fungal virulence was measured with two methods of cuticle (d) and injection (e) infection. Statistical significance between curves was calculated by the log-rank test. Median lethal time (LT50) was calculated by the Kaplan-Meier method, and the values for the cuticle and injection infection methods were shown in panel F and G, respectively. ‘N’ indicates that LT50 value could not be estimated due to the accumulative mortality less than 50%. Statistical significance in phenotype between the WT and gene disruption mutant strain was calculated with Student’s t-test. *: P<0.05; ***: P<0.001; ****: P<0.0001.
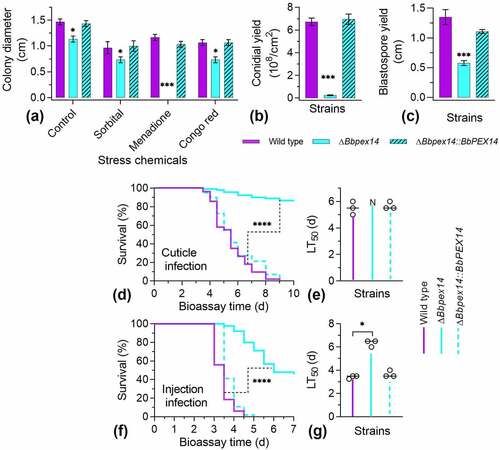
Figure 3. Pex14 significantly is required for the pexophagy process of B. bassiana. Peroxisomes in the wild-type and ΔBbpex14 mutant strains were labeled with the fusion protein BbThi-GFP (PTS2-GFP), and vacuoles were indicated by fluorescence emitted from 7-amino-4-chloromethylcoumarin (CMAC). Fungal strain was grown in SDB medium under submerged condition. The resultant mycelia were subjected to the starvation and oxidative stresses. Without stress, globular and punctuate green signals were obviously observed out of the vacuoles. Under stresses, green signals were translocated into the vacuoles in the wild-type strain, but no significant signals were observed in those of ΔBbpex14 mutant strain. The arrows indicate the signals that appear in the vacuoles. Bars: 5 µm.
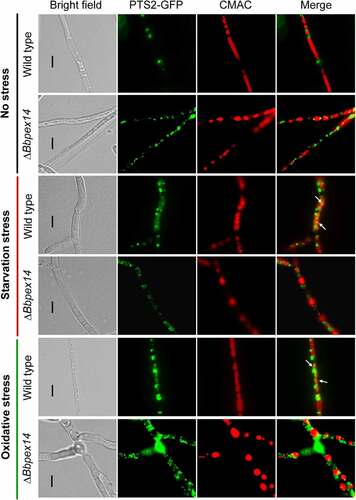
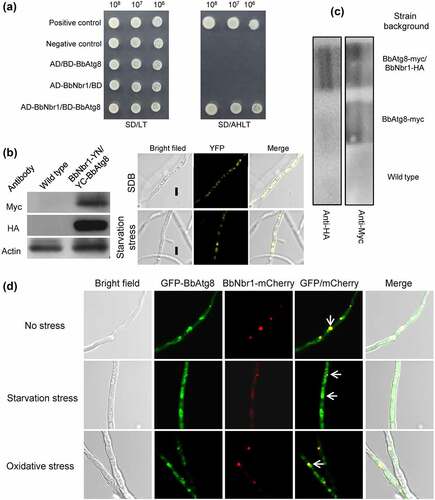
Figure 4. Pex14 interacts with Nbr1 in B. bassiana. (a) Yeast two-hybrid (Y2H) assay. Yeast transformants were initially grown on SD/Leu-Trp (LT) medium. The protein interaction was tested on SD/Ade-His-Leu-Trp (SD/AHLT) and further confirmed on SD/AHLT plus 20 mM 3-amino-1, 2, 4-triazole (AT). Positive and negative controls were provided by kit. (b) Bimolecular fluorescence complementation (BiFC) assay. The N-terminal and C-terminal fragments of yellow fluorescent protein (YFP) (YN and YC) were fused to BbPEX14 and BbNBR1, respectively. The YN and YC of the split YFP contained the coding sequence for myc and HA tag, respectively. Mycelia were cultured in SDB and stressed under starvation. The presence of their proteins was confirmed with western blot. Yellow fluorescence was seen in the mycelial cytoplasm. (c) In co-immunoprecipitation, BbPEX14 and BbNBR1 were fused to Myc and HA, respectively. Protein complex was precipitated with anti-myc magnetic beads, and the presence of BbNbr1 was detected with anti-HA antibody. (d) Co-localization assay. The fusion genes BbPEX14-GFP (representing peroxisomes) and BbNBR1-mCherry were transformed into the wild-type strain. Microscopy imaging revealed that most green signals co-localized with the red signals in the submerged mycelia, and this association persisted in the mycelia under the starvation and oxidative stresses. The arrows indicate the association between the red and green signals. Bar: 5 µm.
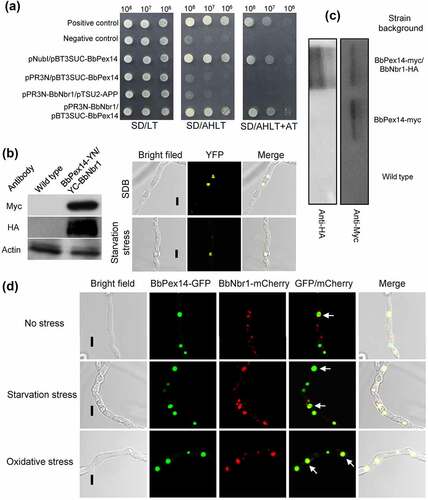
Figure 5. Effects of the BbNbr1 loss on fungal phenotypes. All phenotypic assays were performed as same as those involved in . Phenotypes included stress response (a), conidiation (b), and blastospore formation (c). Insect bioassay was conducted with two infection methods of cuticle application (d) and intra-hemoceol injection (e). Median lethal time (LT50) was calculated by the Kaplan-Meier method, and the values for the cuticle and injection infection methods were shown in panel F and G, respectively. Statistical significance between curves was calculated by the log-rank test, and other phenotypic differences between the WT and ΔBbnbr1 strains were determined with Student’s t-test. *: P<0.05; ***: P<0.001; ****: P<0.0001.
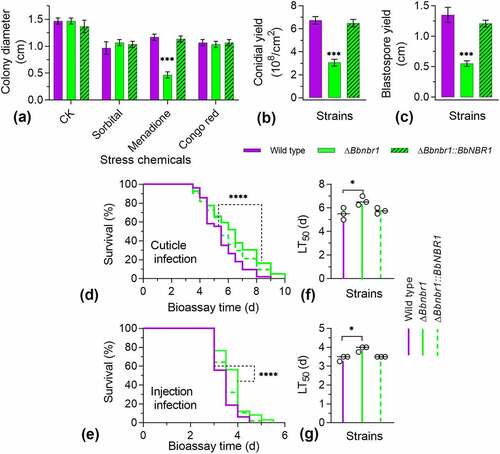
Figure 6. The Nbr1 role involved in the pexophagy process of B. bassiana. Autophagosomes were labeled with the fusion protein GFP-Atg8, and vacuoles were indicated by fluorescence emitted from 7-amino-4-chloromethylcoumarin (CMAC). Mycelia were cultured in SDB medium and exposed the starvation and oxidative stresses. Without stress, globular and punctuate green signals were evenly distributed in cytoplasm. Upon under stresses, green signals appeared in the vacuoles of the wild-type strain, but no significant signals were observed in those of the gene disruption mutant strain. The arrows indicate the signals that appear in the vacuoles. Bars: 5 µm.
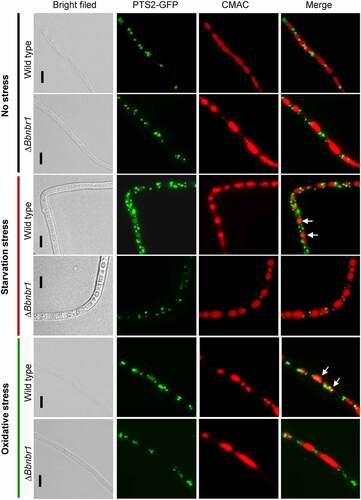
Figure 7. Nbr1 is associated with autophagosomes in B. bassiana. (a) Yeast two-hybrid (Y2H) assay was used to test the interaction between BbAtg8 and BbNbr1. Yeast strains were initially grown on SD/Leu-Trp (LT) medium and confirmed on SD/Ade-His-Leu-Trp (SD/AHLT). Positive and negative controls were provided by kit. Methods for bimolecular fluorescence complementation (b) and co-immunoprecipitation (co-IP) assays (c) were as same as those in . The N-terminal and C-terminal fragments of yellow fluorescent protein (YFP) (YN and YC) were fused to BbNBR1 and BbATG8, respectively. In co-IP, BbATG8 and BbNBR1 were fused to Myc and HA, respectively. (d) In co-localization experiment, the fusion genes GFP-BbATG8 (representing autophagosomes) and BbNBR1-mCherry were transformed into the wild-type strain. Microscopy imaging revealed that the red signals co-localized with the green signals in the stressed and non-stressed mycelia. The arrows indicate the association between the red and green signals. Bar: 5 µm.